Missense variants in ANO4 cause sporadic encephalopathic or familial epilepsy with evidence for a dominant-negative effect
- PMID: 38744284
- PMCID: PMC11179416
- DOI: 10.1016/j.ajhg.2024.04.014
Missense variants in ANO4 cause sporadic encephalopathic or familial epilepsy with evidence for a dominant-negative effect
Abstract
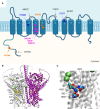
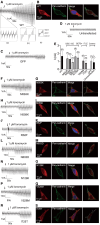
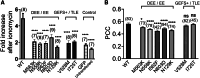
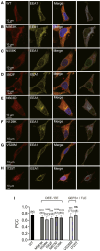
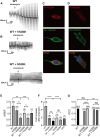
Anoctamins are a family of Ca2+-activated proteins that may act as ion channels and/or phospholipid scramblases with limited understanding of function and disease association. Here, we identified five de novo and two inherited missense variants in ANO4 (alias TMEM16D) as a cause of fever-sensitive developmental and epileptic or epileptic encephalopathy (DEE/EE) and generalized epilepsy with febrile seizures plus (GEFS+) or temporal lobe epilepsy. In silico modeling of the ANO4 structure predicted that all identified variants lead to destabilization of the ANO4 structure. Four variants are localized close to the Ca2+ binding sites of ANO4, suggesting impaired protein function. Variant mapping to the protein topology suggests a preliminary genotype-phenotype correlation. Moreover, the observation of a heterozygous ANO4 deletion in a healthy individual suggests a dysfunctional protein as disease mechanism rather than haploinsufficiency. To test this hypothesis, we examined mutant ANO4 functional properties in a heterologous expression system by patch-clamp recordings, immunocytochemistry, and surface expression of annexin A5 as a measure of phosphatidylserine scramblase activity. All ANO4 variants showed severe loss of ion channel function and DEE/EE associated variants presented mild loss of surface expression due to impaired plasma membrane trafficking. Increased levels of Ca2+-independent annexin A5 at the cell surface suggested an increased apoptosis rate in DEE-mutant expressing cells, but no changes in Ca2+-dependent scramblase activity were observed. Co-transfection with ANO4 wild-type suggested a dominant-negative effect. In summary, we expand the genetic base for both encephalopathic sporadic and inherited fever-sensitive epilepsies and link germline variants in ANO4 to a hereditary disease.
Keywords: ANO4; Ca(2+)-dependent ion channel; GEFS+; TMEM16D; anoctamin; developmental and epileptic encephalopathy; phospholipid scramblase; temporal lobe epilepsy.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests K.H. is currently employed by Janssen Research & Development, Janssen Pharmaceutica N.V., Turnhoutseweg 30, Beerse B-2340, Belgium.
Figures




References
-
- Grether A., Ivanovski I., Russo M., Begemann A., Steindl K., Abela L., Papik M., Zweier M., Oneda B., Joset P., Rauch A. The current benefit of genome sequencing compared to exome sequencing in patients with developmental or epileptic encephalopathies. Mol. Genet. Genomic Med. 2023;11 doi: 10.1002/mgg3.2148. - DOI - PMC - PubMed
-
- Papuc S.M., Abela L., Steindl K., Begemann A., Simmons T.L., Schmitt B., Zweier M., Oneda B., Socher E., Crowther L.M., et al. The role of recessive inheritance in early-onset epileptic encephalopathies: a combined whole-exome sequencing and copy number study. Eur. J. Hum. Genet. 2019;27:408–421. doi: 10.1038/s41431-018-0299-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

