METTL8 links mt-tRNA m3C modification to the HIF1α/RTK/Akt axis to sustain GBM stemness and tumorigenicity
- PMID: 38744809
- PMCID: PMC11093979
- DOI: 10.1038/s41419-024-06718-2
METTL8 links mt-tRNA m3C modification to the HIF1α/RTK/Akt axis to sustain GBM stemness and tumorigenicity
Erratum in
-
Author Correction: METTL8 links mt-tRNA m3C modification to the HIF1α/RTK/Akt axis to sustain GBM stemness and tumorigenicity.Cell Death Dis. 2024 Jul 31;15(7):547. doi: 10.1038/s41419-024-06917-x. Cell Death Dis. 2024. PMID: 39085240 Free PMC article. No abstract available.
Abstract
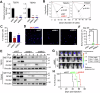
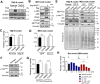
Epitranscriptomic RNA modifications are crucial for the maintenance of glioma stem cells (GSCs), the most malignant cells in glioblastoma (GBM). 3-methylcytosine (m3C) is a new epitranscriptomic mark on RNAs and METTL8 represents an m3C writer that is dysregulated in cancer. Although METTL8 has an established function in mitochondrial tRNA (mt-tRNA) m3C modification, alternative splicing of METTL8 can also generate isoforms that localize to the nucleolus where they may regulate R-loop formation. The molecular basis for METTL8 dysregulation in GBM, and which METTL8 isoform(s) may influence GBM cell fate and malignancy remain elusive. Here, we investigated the role of METTL8 in regulating GBM stemness and tumorigenicity. In GSC, METTL8 is exclusively localized to the mitochondrial matrix where it installs m3C on mt-tRNAThr/Ser(UCN) for mitochondrial translation and respiration. High expression of METTL8 in GBM is attributed to histone variant H2AZ-mediated chromatin accessibility of HIF1α and portends inferior glioma patient outcome. METTL8 depletion impairs the ability of GSC to self-renew and differentiate, thus retarding tumor growth in an intracranial GBM xenograft model. Interestingly, METTL8 depletion decreases protein levels of HIF1α, which serves as a transcription factor for several receptor tyrosine kinase (RTK) genes, in GSC. Accordingly, METTL8 loss inactivates the RTK/Akt axis leading to heightened sensitivity to Akt inhibitor treatment. These mechanistic findings, along with the intimate link between METTL8 levels and the HIF1α/RTK/Akt axis in glioma patients, guided us to propose a HIF1α/Akt inhibitor combination which potently compromises GSC proliferation/self-renewal in vitro. Thus, METTL8 represents a new GBM dependency that is therapeutically targetable.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

