Functional and antigenic characterization of SARS-CoV-2 spike fusion peptide by deep mutational scanning
- PMID: 38744813
- PMCID: PMC11094058
- DOI: 10.1038/s41467-024-48104-8
Functional and antigenic characterization of SARS-CoV-2 spike fusion peptide by deep mutational scanning
Abstract
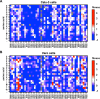
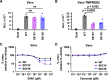
The fusion peptide of SARS-CoV-2 spike protein is functionally important for membrane fusion during virus entry and is part of a broadly neutralizing epitope. However, sequence determinants at the fusion peptide and its adjacent regions for pathogenicity and antigenicity remain elusive. In this study, we perform a series of deep mutational scanning (DMS) experiments on an S2 region spanning the fusion peptide of authentic SARS-CoV-2 in different cell lines and in the presence of broadly neutralizing antibodies. We identify mutations at residue 813 of the spike protein that reduced TMPRSS2-mediated entry with decreased virulence. In addition, we show that an F823Y mutation, present in bat betacoronavirus HKU9 spike protein, confers resistance to broadly neutralizing antibodies. Our findings provide mechanistic insights into SARS-CoV-2 pathogenicity and also highlight a potential challenge in developing broadly protective S2-based coronavirus vaccines.
© 2024. The Author(s).
Conflict of interest statement
N.C.W. consults for HeliXon. The authors declare no competing interests.
Figures



Update of
-
Functional and antigenic characterization of SARS-CoV-2 spike fusion peptide by deep mutational scanning.bioRxiv [Preprint]. 2023 Nov 29:2023.11.28.569051. doi: 10.1101/2023.11.28.569051. bioRxiv. 2023. Update in: Nat Commun. 2024 May 14;15(1):4056. doi: 10.1038/s41467-024-48104-8. PMID: 38076875 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

