Molecular subgroups of T-cell acute lymphoblastic leukemia in adults treated according to pediatric-based GMALL protocols
- PMID: 38744920
- PMCID: PMC11147771
- DOI: 10.1038/s41375-024-02264-0
Molecular subgroups of T-cell acute lymphoblastic leukemia in adults treated according to pediatric-based GMALL protocols
Abstract
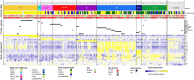
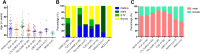
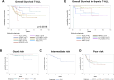
In contrast to B-cell precursor acute lymphoblastic leukemia (ALL), molecular subgroups are less well defined in T-lineage ALL. Comprehensive studies on molecular T-ALL subgroups have been predominantly performed in pediatric ALL patients. Currently, molecular characteristics are rarely considered for risk stratification. Herein, we present a homogenously treated cohort of 230 adult T-ALL patients characterized on transcriptome, and partly on DNA methylation and gene mutation level in correlation with clinical outcome. We identified nine molecular subgroups based on aberrant oncogene expression correlating to four distinct DNA methylation patterns. The subgroup distribution differed from reported pediatric T-ALL cohorts with higher frequencies of prognostic unfavorable subgroups like HOXA or LYL1/LMO2. A small subset (3%) of HOXA adult T-ALL patients revealed restricted expression of posterior HOX genes with aberrant activation of lncRNA HOTTIP. With respect to outcome, TLX1 (n = 44) and NKX2-1 (n = 4) had an exceptionally favorable 3-year overall survival (3y-OS) of 94%. Within thymic T-ALL, the non TLX1 patients had an inferior but still good prognosis. To our knowledge this is the largest cohort of adult T-ALL patients characterized by transcriptome sequencing with meaningful clinical follow-up. Risk classification based on molecular subgroups might emerge and contribute to improvements in outcome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bassan R, Bourquin JP, DeAngelo DJ, Chiaretti S. New approaches to the management of adult acute lymphoblastic leukemia. J Clin Oncol. 2018;36:JCO2017773648. - PubMed
-
- Goekbuget N, Fiedler W, Alakel N, Topp MS, Hanoun M, Steffen B, et al. Results of the risk-adapted, MRD-stratified GMALL trial 08/2013 in 281 T-ALL / T-LLB patients: excellent outcome of standard risk thymic T-ALL. Blood. 2022;140:115–7. doi: 10.1182/blood-2022-158381. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

