Inseparable RNA binding and chromatin modification activities of a nucleosome-interacting surface in EZH2
- PMID: 38744974
- PMCID: PMC11176075
- DOI: 10.1038/s41588-024-01740-8
Inseparable RNA binding and chromatin modification activities of a nucleosome-interacting surface in EZH2
Abstract
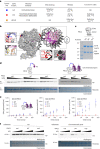
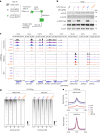
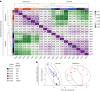
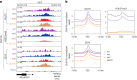
Polycomb repressive complex 2 (PRC2) interacts with RNA in cells, but there is no consensus on how RNA regulates PRC2 canonical functions, including chromatin modification and the maintenance of transcription programs in lineage-committed cells. We assayed two separation-of-function mutants of the PRC2 catalytic subunit EZH2, defective in RNA binding but functional in methyltransferase activity. We find that part of the RNA-binding surface of EZH2 is required for chromatin modification, yet this activity is independent of RNA. Mechanistically, the RNA-binding surface within EZH2 is required for chromatin modification in vitro and in cells, through interactions with nucleosomal DNA. Contrarily, an RNA-binding-defective mutant exhibited normal chromatin modification activity in vitro and in lineage-committed cells, accompanied by normal gene repression activity. Collectively, we show that part of the RNA-binding surface of EZH2, rather than the RNA-binding activity per se, is required for the histone methylation in vitro and in cells, through interactions with the substrate nucleosome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Bhat P, Honson D, Guttman M. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Rev. Mol. Cell Biol. 2021;22:653–670. - PubMed
-
- Cech TR, Steitz JA. The noncoding RNA revolution–trashing old rules to forge new ones. Cell. 2014;157:77–94. - PubMed
-
- Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. - PubMed
MeSH terms
Substances
Grants and funding
- DP190103407/Department of Education and Training | Australian Research Council (ARC)
- APP1162921/Department of Health | National Health and Medical Research Council (NHMRC)
- APP1184637/Department of Health | National Health and Medical Research Council (NHMRC)
- APP2011767/Department of Health | National Health and Medical Research Council (NHMRC)
- APP2020900/Department of Health | National Health and Medical Research Council (NHMRC)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

