Mutant p53 reactivation restricts the protumorigenic consequences of wild type p53 loss of heterozygosity in Li-Fraumeni syndrome patient-derived fibroblasts
- PMID: 38745079
- PMCID: PMC11239894
- DOI: 10.1038/s41418-024-01307-4
Mutant p53 reactivation restricts the protumorigenic consequences of wild type p53 loss of heterozygosity in Li-Fraumeni syndrome patient-derived fibroblasts
Abstract
The p53 tumor suppressor, encoded by the TP53 gene, serves as a major barrier against malignant transformation. Patients with Li-Fraumeni syndrome (LFS) inherit a mutated TP53 allele from one parent and a wild-type TP53 allele from the other. Subsequently, the wild-type allele is lost and only the mutant TP53 allele remains. This process, which is termed loss of heterozygosity (LOH), results in only mutant p53 protein expression. We used primary dermal fibroblasts from LFS patients carrying the hotspot p53 gain-of-function pathogenic variant, R248Q to study the LOH process and characterize alterations in various pathways before and after LOH. We previously described the derivation of mutant p53 reactivating peptides, designated pCAPs (p53 Conformation Activating Peptides). In this study, we tested the effect of lead peptide pCAP-250 on LOH and on its associated cellular changes. We report that treatment of LFS fibroblasts with pCAP-250 prevents the accumulation of mutant p53 protein, inhibits LOH, and alleviates its cellular consequences. Furthermore, prolonged treatment with pCAP-250 significantly reduces DNA damage and restores long-term genomic stability. pCAPs may thus be contemplated as a potential preventive treatment to prevent or delay early onset cancer in carriers of mutant p53.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
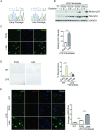

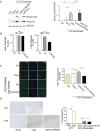
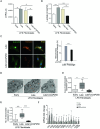


References
-
- Bártek J, Bártková J, Vojtĕsek B, Stasková Z, Lukás J, Rejthar A, et al. Aberrant expression of the p53 oncoprotein is a common feature of a wide spectrum of human malignancies. Oncogene. 1991;6:1699–703. - PubMed
-
- Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–78. - PubMed
-
- Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 Protein in the Cellular Response to DNA Damage. Cancer Res. 1991;51:6304–11. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

