This is a preprint.
Divergent gene expression patterns in alcohol and opioid use disorders lead to consistent alterations in functional networks within the Dorsolateral Prefrontal Cortex
- PMID: 38746311
- PMCID: PMC11092658
- DOI: 10.1101/2024.04.29.591734
Divergent gene expression patterns in alcohol and opioid use disorders lead to consistent alterations in functional networks within the Dorsolateral Prefrontal Cortex
Update in
-
Divergent gene expression patterns in alcohol and opioid use disorders lead to consistent alterations in functional networks within the dorsolateral prefrontal cortex.Transl Psychiatry. 2024 Oct 14;14(1):437. doi: 10.1038/s41398-024-03143-z. Transl Psychiatry. 2024. PMID: 39402051 Free PMC article.
Abstract
Substance Use Disorders (SUDs) manifest as persistent drug-seeking behavior despite adverse consequences, with Alcohol Use Disorder (AUD) and Opioid Use Disorder (OUD) representing prevalent forms associated with significant mortality rates and economic burdens. The co-occurrence of AUD and OUD is common, necessitating a deeper comprehension of their intricate interactions. While the causal link between these disorders remains elusive, shared genetic factors are hypothesized. Leveraging public datasets, we employed genomic and transcriptomic analyses to explore conserved and distinct molecular pathways within the dorsolateral prefrontal cortex associated with AUD and OUD. Our findings unveil modest transcriptomic overlap at the gene level between the two disorders but substantial convergence on shared biological pathways. Notably, these pathways predominantly involve inflammatory processes, synaptic plasticity, and key intracellular signaling regulators. Integration of transcriptomic data with the latest genome-wide association studies (GWAS) for problematic alcohol use (PAU) and OUD not only corroborated our transcriptomic findings but also confirmed the limited shared heritability between the disorders. Overall, our study indicates that while alcohol and opioids induce diverse transcriptional alterations at the gene level, they converge on select biological pathways, offering promising avenues for novel therapeutic targets aimed at addressing both disorders simultaneously.
Figures




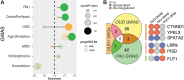
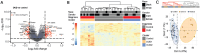

References
-
- Deaths from Excessive Alcohol Use in the United States. Centers for Disease Control and Prevention. Feb. 29, 2024. URL: https://www.cdc.gov/alcohol/features/excessive-alcohol-deaths.html (visited on 04/09/2024).
-
- Drug Overdose Deaths. Centers for Disease Control and Prevention. Aug. 22, 2023. URL: https://www.cdc.gov/drugoverdose/deaths/index.html (visited on 04/09/2024).
-
- Excessive Drinking is Draining the U.S. Economy. Centers for Disease Control and Prevention. Feb. 23, 2024. URL: https://www.cdc.gov/alcohol/features/excessive-drinking.html (visited on 04/09/2024).
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
