MitoH3: Mitochondrial Haplogroup and Homoplasmic/Heteroplasmic Variant Calling Pipeline for Alzheimer's Disease Sequencing Project
- PMID: 38746629
- PMCID: PMC11091720
- DOI: 10.3233/ADR-230120
MitoH3: Mitochondrial Haplogroup and Homoplasmic/Heteroplasmic Variant Calling Pipeline for Alzheimer's Disease Sequencing Project
Abstract
Background: Mitochondrial DNA (mtDNA) is a double-stranded circular DNA and has multiple copies in each cell. Excess heteroplasmy, the coexistence of distinct variants in copies of mtDNA within a cell, may lead to mitochondrial impairments. Accurate determination of heteroplasmy in whole-genome sequencing (WGS) data has posed a significant challenge because mitochondria carrying heteroplasmic variants cannot be distinguished during library preparation. Moreover, sequencing errors, contamination, and nuclear mtDNA segments can reduce the accuracy of heteroplasmic variant calling.
Objective: To efficiently and accurately call mtDNA homoplasmic and heteroplasmic variants from the large-scale WGS data generated from the Alzheimer's Disease Sequencing Project (ADSP), and test their association with Alzheimer's disease (AD).
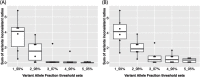
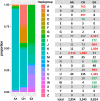
Methods: In this study, we present MitoH3-a comprehensive computational pipeline for calling mtDNA homoplasmic and heteroplasmic variants and inferring haplogroups in the ADSP WGS data. We first applied MitoH3 to 45 technical replicates from 6 subjects to define a threshold for detecting heteroplasmic variants. Then using the threshold of 5% ≤variant allele fraction≤95%, we further applied MitoH3 to call heteroplasmic variants from a total of 16,113 DNA samples with 6,742 samples from cognitively normal controls and 6,183 from AD cases.
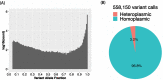
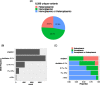
Results: This pipeline is available through the Singularity container engine. For 4,311 heteroplasmic variants identified from 16,113 samples, no significant variant count difference was observed between AD cases and controls.
Conclusions: Our streamlined pipeline, MitoH3, enables computationally efficient and accurate analysis of a large number of samples.
Keywords: Alzheimer’s disease; haplogroup; homoplasmic and heteroplasmic variant calling; mitochondrial DNA; whole genome sequencing.
© 2024 – The authors. Published by IOS Press.
Conflict of interest statement
The authors have no conflict of interest to report.
Figures
References
-
- Stewart JB, Chinnery PF (2015) The dynamics of mitochondrial DNA heteroplasmy: Implications for human health and disease. Nat Rev Genet 16, 530–542. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

