In silico evaluation of anti-colorectal cancer inhibitors by Resveratrol derivatives targeting Armadillo repeats domain of APC: molecular docking and molecular dynamics simulation
- PMID: 38746675
- PMCID: PMC11091374
- DOI: 10.3389/fonc.2024.1360745
In silico evaluation of anti-colorectal cancer inhibitors by Resveratrol derivatives targeting Armadillo repeats domain of APC: molecular docking and molecular dynamics simulation
Abstract
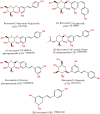
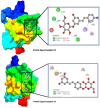
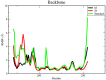
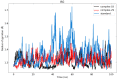
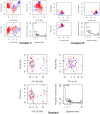
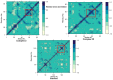
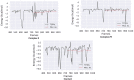
Colorectal cancer is the second leading cause of cancer-related deaths. In 2018, there were an estimated 1.8 million cases, and this number is expected to increase to 2.2 million by 2030. Despite its prevalence, the current therapeutic option has a lot of side effects and limitations. Therefore, this study was designed to employ a computational approach for the identification of anti-cancer inhibitors against colorectal cancer using Resveratrol derivatives. Initially, the pass prediction spectrum of 50 derivatives was conducted and selected top seven compounds based on the maximum pass prediction score. After that, a comprehensive analysis, including Lipinski Rule, pharmacokinetics, ADMET profile study, molecular orbitals analysis, molecular docking, molecular dynamic simulations, and MM-PBSA binding free energy calculations. The reported binding affinity ranges of Resveratrol derivatives from molecular docking were -6.1 kcal/mol to -7.9 kcal/mol against the targeted receptor of human armadillo repeats domain of adenomatous polyposis coli (APC) (PDB ID: 3NMW). Specifically, our findings reported that two compounds [(03) Resveratrol 3-beta-mono-D-glucoside, and (29) Resveratrol 3-Glucoside] displayed the highest level of effectiveness compared to all other derivatives (-7.7 kcal/mol and -7.9 kcal/mol), and favorable drug-likeness, and exceptional safety profiles. Importantly, almost all the molecules were reported as free from toxic effects. Subsequently, molecular dynamic simulations conducted over 100ns confirmed the stability of the top two ligand-protein complexes. These findings suggest that Resveratrol derivatives may be effective drug candidate to manage the colorectal cancer. However, further experimental research, such as in vitro/in vivo studies, is essential to validate these computational findings and confirm their practical value.
Keywords: colorectal cancer; drug design; molecular docking; molecular dynamics simulation; resveratrol derivatives.
Copyright © 2024 Akash, Islam, Bhuiyan, Islam, Bayıl, Saleem, Albadrani, Al-Ghadi and Abdel-Daim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Investigation of the New Inhibitors by Sulfadiazine and Modified Derivatives of α-D-glucopyranoside for White Spot Syndrome Virus Disease of Shrimp by In Silico: Quantum Calculations, Molecular Docking, ADMET and Molecular Dynamics Study.Molecules. 2022 Jun 8;27(12):3694. doi: 10.3390/molecules27123694. Molecules. 2022. PMID: 35744817 Free PMC article.
-
In silico design of novel bioactive molecules to treat breast cancer with chlorogenic acid derivatives: a computational and SAR approach.Front Pharmacol. 2023 Dec 12;14:1266833. doi: 10.3389/fphar.2023.1266833. eCollection 2023. Front Pharmacol. 2023. PMID: 38152692 Free PMC article.
-
A drug design strategy based on molecular docking and molecular dynamics simulations applied to development of inhibitor against triple-negative breast cancer by Scutellarein derivatives.PLoS One. 2023 Oct 12;18(10):e0283271. doi: 10.1371/journal.pone.0283271. eCollection 2023. PLoS One. 2023. PMID: 37824496 Free PMC article.
-
Identification of novel PI3Kδ inhibitors by docking, ADMET prediction and molecular dynamics simulations.Comput Biol Chem. 2019 Feb;78:190-204. doi: 10.1016/j.compbiolchem.2018.12.002. Epub 2018 Dec 7. Comput Biol Chem. 2019. PMID: 30557817
-
Identification of Kaempferol as Viral Entry Inhibitor and DL-Arginine as Viral Replication Inhibitor from Selected Plants of Indian Traditional Medicine against COVID-19: An in silico Guided in vitro Approach.Curr Comput Aided Drug Des. 2023;19(4):313-323. doi: 10.2174/1573409919666230112123213. Curr Comput Aided Drug Des. 2023. PMID: 36635906
Cited by
-
In-silico discovery of type-2 diabetes-causing host key genes that are associated with the complexity of monkeypox and repurposing common drugs.Brief Bioinform. 2025 May 1;26(3):bbaf215. doi: 10.1093/bib/bbaf215. Brief Bioinform. 2025. PMID: 40370100 Free PMC article.
-
In silico identification of PPARγ agonists from diffractaic acid analogs in prostate cancer: a comprehensive computational approach.3 Biotech. 2025 Jul;15(7):219. doi: 10.1007/s13205-025-04376-5. Epub 2025 Jun 20. 3 Biotech. 2025. PMID: 40546399
-
Exploring common genomic biomarkers to disclose common drugs for the treatment of colorectal cancer and hepatocellular carcinoma with type-2 diabetes through transcriptomics analysis.PLoS One. 2025 Mar 24;20(3):e0319028. doi: 10.1371/journal.pone.0319028. eCollection 2025. PLoS One. 2025. PMID: 40127075 Free PMC article.
-
Structure-based virtual screening of Trachyspermum ammi metabolites targeting acetylcholinesterase for Alzheimer's disease treatment.PLoS One. 2024 Dec 17;19(12):e0311401. doi: 10.1371/journal.pone.0311401. eCollection 2024. PLoS One. 2024. Retraction in: PLoS One. 2025 May 8;20(5):e0324094. doi: 10.1371/journal.pone.0324094. PMID: 39689077 Free PMC article. Retracted.
References
-
- Pitchumoni C, Broder A. Epidemiology of colorectal cancer. In: Colorectal Neoplasia and the Colorectal Microbiome. Elsevier; (2020). p. 5–33.
-
- Roper J, Hung KE. Molecular mechanisms of colorectal carcinogenesis. Mol Pathogene Colorectal Cancer. (2013) 25-65.
LinkOut - more resources
Full Text Sources

