Integrated Hfq-interacting RNAome and transcriptomic analysis reveals complex regulatory networks of nitrogen fixation in root-associated Pseudomonas stutzeri A1501
- PMID: 38747590
- PMCID: PMC11332353
- DOI: 10.1128/msphere.00762-23
Integrated Hfq-interacting RNAome and transcriptomic analysis reveals complex regulatory networks of nitrogen fixation in root-associated Pseudomonas stutzeri A1501
Abstract
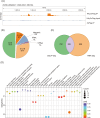
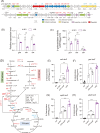
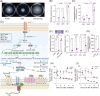
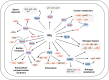
The RNA chaperone Hfq acts as a global regulator of numerous biological processes, such as carbon/nitrogen metabolism and environmental adaptation in plant-associated diazotrophs; however, its target RNAs and the mechanisms underlying nitrogen fixation remain largely unknown. Here, we used enhanced UV cross-linking immunoprecipitation coupled with high-throughput sequencing to identify hundreds of Hfq-binding RNAs probably involved in nitrogen fixation, carbon substrate utilization, biofilm formation, and other functions. Collectively, these processes endow strain A1501 with the requisite capabilities to thrive in the highly competitive rhizosphere. Our findings revealed a previously uncharted landscape of Hfq target genes. Notable among these is nifM, encoding an isomerase necessary for nitrogenase reductase solubility; amtB, encoding an ammonium transporter; oprB, encoding a carbohydrate porin; and cheZ, encoding a chemotaxis protein. Furthermore, we identified more than 100 genes of unknown function, which expands the potential direct regulatory targets of Hfq in diazotrophs. Our data showed that Hfq directly interacts with the mRNA of regulatory proteins (RsmA, AlgU, and NifA), regulatory ncRNA RsmY, and other potential targets, thus revealing the mechanistic links in nitrogen fixation and other metabolic pathways.
Importance: Numerous experimental approaches often face challenges in distinguishing between direct and indirect effects of Hfq-mediated regulation. New technologies based on high-throughput sequencing are increasingly providing insight into the global regulation of Hfq in gene expression. Here, enhanced UV cross-linking immunoprecipitation coupled with high-throughput sequencing was employed to identify the Hfq-binding sites and potential targets in the root-associated Pseudomonas stutzeri A1501 and identify hundreds of novel Hfq-binding RNAs that are predicted to be involved in metabolism, environmental adaptation, and nitrogen fixation. In particular, we have shown Hfq interactions with various regulatory proteins' mRNA and their potential targets at the posttranscriptional level. This study not only enhances our understanding of Hfq regulation but, importantly, also provides a framework for addressing integrated regulatory network underlying root-associated nitrogen fixation.
Keywords: Hfq; Pseudomonas stutzeri; eCLIP-seq; nitrogen fixation; posttranscriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Zhang H, Zhan Y, Yan Y, Liu Y, Hu G, Wang S, Yang H, Qiu X, Liu Y, Li J, Lu W, Elmerich C, Lin M. 2019. The Pseudomonas stutzeri-specific regulatory noncoding RNA Nfis targets katB mRNA encoding a catalase essential for optimal oxidative resistance and nitrogenase activity. J Bacteriol 201:1–16. doi:10.1128/JB.00334-19 - DOI - PMC - PubMed
-
- Moreno R, Hernández-Arranz S, La Rosa R, Yuste L, Madhushani A, Shingler V, Rojo F. 2015. The Crc and Hfq proteins of Pseudomonas putida cooperate in catabolite repression and formation of ribonucleic acid complexes with specific target motifs. Environ Microbiol 17:105–118. doi:10.1111/1462-2920.12499 - DOI - PubMed
-
- Lv F, Zhan Y, Lu W, Ke X, Shao Y, Ma Y, Zheng J, Yang Z, Jiang S, Shang L, Ma Y, Cheng L, Elmerich C, Yan Y, Lin M. 2022. Regulation of hierarchical carbon substrate utilization, nitrogen fixation and root colonization by the Hfq/Crc/Crczy genes in Pseudomonas stutzeri. iScience 25:105663. doi:10.1016/j.isci.2022.105663 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFA0912100/MOST | National Key Research and Development Program of China (NKPs)
- 2019YFA0904700/MOST | National Key Research and Development Program of China (NKPs)
- 31930004/MOST | National Natural Science Foundation of China (NSFC)
- 32150021/MOST | National Natural Science Foundation of China (NSFC)
- 32270067/MOST | National Natural Science Foundation of China (NSFC)
LinkOut - more resources
Full Text Sources
