Impact of whole grain highland hull-less barley on the denaturing gradient gel electrophoresis profiles of gut microbial communities in rats fed high-fat diets
- PMID: 38747621
- PMCID: PMC11237377
- DOI: 10.1128/spectrum.04089-23
Impact of whole grain highland hull-less barley on the denaturing gradient gel electrophoresis profiles of gut microbial communities in rats fed high-fat diets
Abstract
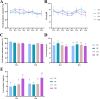
Polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) is a traditional non-culture technique that can provide a fingerprint of the microbial community. In the field of gut microbiota analysis, PCR-DGGE still holds potential for development. In the present study, we utilized an improved nested PCR-DGGE approach targeting the V3 region of 16S ribosomal DNA to investigate the impact of whole grain highland hull-less barley (WHLB), a cereal known for its significant hypocholesterolemic effect, on the gut microbiota profiles of high-fat diet rats. Seventy-two male Sprague-Dawley rats were divided into four groups and fed a normal control diet, a high-fat diet, or a high-fat diet supplemented with a low or high dose of WHLB for 4 or 8 weeks. The results revealed that the dominant bands varied among different dose groups and further changed with different treatment times. The compositions of bacterial communities in feces and cecal content were similar, but the dominant bacterial bands differed. After performing double DGGE, extracting the bands, sequencing the DNA, and aligning the sequences, a total of 19 bands were classified under the Firmicutes and Bacteroidetes phyla, while two bands were identified as unclassified uncultured bacteria. The relative abundance of Lactobacillus gasseri, Uncultured Prevotella sp., and Clostridium sp. increased following the administration of WHLB. Illumina-based sequencing was employed to assess the reliability of DGGE, demonstrating its reliability in analyzing the dominant taxonomic composition, although it may have limitations in accurately detecting the alpha diversity of bacterial species.
Importance: While next-generation sequencing has overshadowed polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE), the latter still holds promise for advancing gut microbiota analysis due to its unique advantages. In this study, we used optimized nested PCR-DGGE to investigate the gut microbiota profile of high-fat diet rats after administering whole grain highland hull-less barley. High-throughput sequencing was employed to validate the DGGE results. Our results proved the reliability of PCR-DGGE for analyzing the dominant taxonomic composition while also providing visual evidence of a notable relationship between the composition of cecal and fecal microbial communities, highlighting substantial differences in both richness and abundance.
Keywords: denaturing gradient gel electrophoresis; fecal microbiota; high-throughput sequencing; nested PCR; taxonomic composition; whole grain.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- WHO . 2021. Cardiovascular diseases. Available from: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases.... Retrieved 9 Jan 2023.
-
- Lan Y, Ning K, Ma Y, Zhao J, Ci C, Yang X, An F, Zhang Z, An Y, Cheng M. 2022. High-density lipoprotein cholesterol as a potential medium between depletion of Lachnospiraceae genera and hypertension under a high-calorie diet. Microbiol Spectr 10:e0234922. doi: 10.1128/spectrum.02349-22 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

