Comparative analysis of SARS-CoV-2 neutralization titers reveals consistency between human and animal model serum and across assays
- PMID: 38748773
- PMCID: PMC12362660
- DOI: 10.1126/scitranslmed.adl1722
Comparative analysis of SARS-CoV-2 neutralization titers reveals consistency between human and animal model serum and across assays
Abstract
The evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) requires ongoing monitoring to judge the ability of newly arising variants to escape the immune response. A surveillance system necessitates an understanding of differences in neutralization titers measured in different assays and using human and animal serum samples. We compared 18 datasets generated using human, hamster, and mouse serum and six different neutralization assays. Datasets using animal model serum samples showed higher titer magnitudes than datasets using human serum samples in this comparison. Fold change in neutralization of variants compared to ancestral SARS-CoV-2, immunodominance patterns, and antigenic maps were similar among serum samples and assays. Most assays yielded consistent results, except for differences in fold change in cytopathic effect assays. Hamster serum samples were a consistent surrogate for human first-infection serum samples. These results inform the transition of surveillance of SARS-CoV-2 antigenic variation from dependence on human first-infection serum samples to the utilization of serum samples from animal models.
Conflict of interest statement
Competing interests
VMC is a co-inventor on a patent application entitled “Methods and reagents for diagnosis of SARS-CoV-2 infection” (Patent application no EP3809137A1). MSD is a consultant for Inbios, Vir Biotechnology, Ocugen, Topspin Therapeutics, Moderna, and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, Generate Biomedicines, and Emergent BioSolutions. YK received unrelated funding support from Daiichi Sankyo Pharmaceutical, Toyama Chemical, Tauns Laboratories, Inc., Shionogi & Co. LTD, Otsuka Pharmaceutical, KM Biologics, Kyoritsu Seiyaku, Shinya Corporation, and Fuji Rebio. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines which list FK as co-inventor: “Influenza virus vaccination regimens” (Patent no 20190125859), “Influenza virus vaccines and uses thereof” (Patent no 9371366), “Influenza virus vaccines and uses thereof” (Patent nos 20180333479, 9968670, 20190099484, 20150335729, 10131695, 10137189, 20140328875, 20160361408, 20150132330, 20190106461, WO2013043729A1 WIPO (PCT), WO2016205347A1 WIPO (PCT)), “Influenza virus hemagglutinin proteins and uses thereof” (Patent nos WO2017218624A1 WIPO (PCT)), “Influenza virus vaccination regimens” (WO2016118937A1 WIPO (PCT)). Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. FK has consulted for Merck, Seqirus, Curevac and Pfizer, and is currently consulting for GSK, Gritstone, 3rd Rock Ventures and Avimex. FK is a co-founder and scientific advisory board member of CastleVax. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2 and Dynavax on influenza virus vaccines. IE has received a research grant and speakers fees from Moderna. B. Meyer has received a research grant from Moderna. GRS is on the GSK Vaccines Scientific Advisory Board. Oxford University holds intellectual property related to the Oxford-AstraZeneca vaccine. MSS serves in an advisory role for Ocugen, Inc. SP reports that the Uniformed Services University (USU) Infectious Diseases Clinical Research Program (IDCRP), a US Department of Defense institution, and the Henry M. Jackson Foundation (HJF) were funded under a Cooperative Research and Development Agreement to conduct an unrelated phase III COVID-19 monoclonal antibody immunoprophylaxis trial sponsored by AstraZeneca. The HJF, in support of the USU IDCRP, was funded by the Department of Defense Joint Program Executive Office for Chemical, Biological, Radiological, and Nuclear Defense to augment the conduct of an unrelated phase III vaccine trial sponsored by AstraZeneca. Both trials were part of the U.S. Government COVID-19 response. Neither is related to the work presented here. Authors not listed above declare that they have no competing interests. The views expressed are those of the authors and do not reflect the official policy of the USUHS, Department of the Army, Department of the Navy, the Department of the Air Force, the Department of Defense or the U.S. Government and the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF). The investigators have adhered to the policies for protection of human subjects as prescribed in 45 CFR 46. SL, WW, and CDW are US Government employees. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by an employee of the U.S. Government as part of that person’s official duties.
Figures
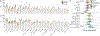


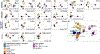
References
-
- Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB, Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 184, 2523 (2021). - PMC - PubMed
-
- Lucas C, Vogels CBF, Yildirim I, Rothman JE, Lu P, Monteiro V, Gehlhausen JR, Campbell M, Silva J, Tabachnikova A, Peña-Hernandez MA, Muenker MC, Breban MI, Fauver JR, Mohanty S, Huang J, Yale SARS-CoV-2 Genomic Surveillance Initiative, Shaw AC, Ko AI, Omer SB, Grubaugh ND, Iwasaki A, Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity. Nature. 600, 523–529 (2021). - PMC - PubMed
-
- Mykytyn AZ, Rissmann M, Kok A, Rosu ME, Schipper D, Breugem TI, van den Doel PB, Chandler F, Bestebroer T, de Wit M, van Royen ME, Molenkamp R, Munnink BBO, de Vries RD, GeurtsvanKessel C, Smith DJ, Koopmans MPG, Rockx B, Lamers MM, Fouchier R, Haagmans BL, Antigenic cartography of SARS-CoV-2 reveals that Omicron BA.1 and BA.2 are antigenically distinct. Science Immunology. 0, eabq4450. - PMC - PubMed
-
- Dejnirattisai W, Huo J, Zhou D, Zahradník J, Supasa P, Liu C, Duyvesteyn HME, Ginn HM, Mentzer AJ, Tuekprakhon A, Nutalai R, Wang B, Dijokaite A, Khan S, Avinoam O, Bahar M, Skelly D, Adele S, Johnson SA, Amini A, Ritter TG, Mason C, Dold C, Pan D, Assadi S, Bellass A, Omo-Dare N, Koeckerling D, Flaxman A, Jenkin D, Aley PK, Voysey M, Costa Clemens SA, Naveca FG, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Baillie V, Serafin N, Kwatra G, Da Silva K, Madhi SA, Nunes MC, Malik T, Openshaw PJM, Baillie JK, Semple MG, Townsend AR, Huang K-YA, Tan TK, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Constantinides B, Webster H, Crook D, Pollard AJ, Lambe T, OPTIC Consortium, ISARIC4C Consortium, Paterson NG, Williams MA, Hall DR, Fry EE, Mongkolsapaya J, Ren J, Schreiber G, Stuart DI, Screaton GR, SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 185, 467–484.e15 (2022). - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

