SARS-CoV-2 infection exacerbates the cellular pathology of Parkinson's disease in human dopaminergic neurons and a mouse model
- PMID: 38749422
- PMCID: PMC11148862
- DOI: 10.1016/j.xcrm.2024.101570
SARS-CoV-2 infection exacerbates the cellular pathology of Parkinson's disease in human dopaminergic neurons and a mouse model
Abstract
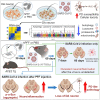
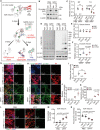
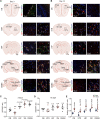
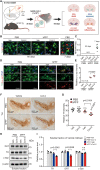
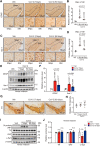
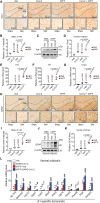
While an association between Parkinson's disease (PD) and viral infections has been recognized, the impact of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) on PD progression remains unclear. Here, we demonstrate that SARS-CoV-2 infection heightens the risk of PD using human embryonic stem cell (hESC)-derived dopaminergic (DA) neurons and a human angiotensin-converting enzyme 2 (hACE2) transgenic (Tg) mouse model. Our findings reveal that SARS-CoV-2 infection exacerbates PD susceptibility and cellular toxicity in DA neurons pre-treated with human preformed fibrils (hPFFs). Additionally, nasally delivered SARS-CoV-2 infects DA neurons in hACE2 Tg mice, aggravating the damage initiated by hPFFs. Mice infected with SARS-CoV-2 display persisting neuroinflammation even after the virus is no longer detectable in the brain. A comprehensive analysis suggests that the inflammatory response mediated by astrocytes and microglia could contribute to increased PD susceptibility associated with SARS-CoV-2. These findings advance our understanding of the potential long-term effects of SARS-CoV-2 infection on the progression of PD.
Keywords: COVID-19 sequalae; DA neuron; PD; Parkinson’s disease; SARS-CoV-2; disease modeling; dopaminergic neuron; hACE2 transgenic mouse; neuroinflammation; neurological sequelae.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Benameur K., Agarwal A., Auld S.C., Butters M.P., Webster A.S., Ozturk T., Howell J.C., Bassit L.C., Velasquez A., Schinazi R.F., et al. Encephalopathy and Encephalitis Associated with Cerebrospinal Fluid Cytokine Alterations and Coronavirus Disease, Atlanta, Georgia, USA, 2020. Emerg. Infect. Dis. 2020;26:2016–2021. doi: 10.3201/eid2609.202122. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

