Graph Artificial Intelligence in Medicine
- PMID: 38749465
- PMCID: PMC11344018
- DOI: 10.1146/annurev-biodatasci-110723-024625
Graph Artificial Intelligence in Medicine
Abstract
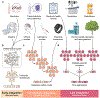
In clinical artificial intelligence (AI), graph representation learning, mainly through graph neural networks and graph transformer architectures, stands out for its capability to capture intricate relationships and structures within clinical datasets. With diverse data-from patient records to imaging-graph AI models process data holistically by viewing modalities and entities within them as nodes interconnected by their relationships. Graph AI facilitates model transfer across clinical tasks, enabling models to generalize across patient populations without additional parameters and with minimal to no retraining. However, the importance of human-centered design and model interpretability in clinical decision-making cannot be overstated. Since graph AI models capture information through localized neural transformations defined on relational datasets, they offer both an opportunity and a challenge in elucidating model rationale. Knowledge graphs can enhance interpretability by aligning model-driven insights with medical knowledge. Emerging graph AI models integrate diverse data modalities through pretraining, facilitate interactive feedback loops, and foster human-AI collaboration, paving the way toward clinically meaningful predictions.
Keywords: artificial intelligence; graph neural networks; graph transformers; health care; human-centered AI; knowledge graphs; medicine; multimodal learning; transfer learning.
Figures
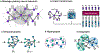

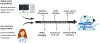
References
-
- Council N. 2011. Committee on a framework for developing a new taxonomy of disease. toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease - PubMed
-
- Data MC, Nair S, Hsu D, Celi LA. 2016. Challenges and opportunities in secondary analyses of electronic health record data. Secondary analysis of electronic health records :17–26 - PubMed
-
- Sandhu E, Weinstein S, McKethan A, Jain SH. 2012. Secondary uses of electronic health record data: benefits and barriers. Joint Commission journal on quality and patient safety 38(1):34–40 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

