Imprinting of serum neutralizing antibodies by Wuhan-1 mRNA vaccines
- PMID: 38749479
- PMCID: PMC11419699
- DOI: 10.1038/s41586-024-07539-1
Imprinting of serum neutralizing antibodies by Wuhan-1 mRNA vaccines
Abstract
Immune imprinting is a phenomenon in which prior antigenic experiences influence responses to subsequent infection or vaccination1,2. The effects of immune imprinting on serum antibody responses after boosting with variant-matched SARS-CoV-2 vaccines remain uncertain. Here we characterized the serum antibody responses after mRNA vaccine boosting of mice and human clinical trial participants. In mice, a single dose of a preclinical version of mRNA-1273 vaccine encoding Wuhan-1 spike protein minimally imprinted serum responses elicited by Omicron boosters, enabling generation of type-specific antibodies. However, imprinting was observed in mice receiving an Omicron booster after two priming doses of mRNA-1273, an effect that was mitigated by a second booster dose of Omicron vaccine. In both SARS-CoV-2-infected and uninfected humans who received two Omicron-matched boosters after two or more doses of the prototype mRNA-1273 vaccine, spike-binding and neutralizing serum antibodies cross-reacted with Omicron variants as well as more distantly related sarbecoviruses. Because serum neutralizing responses against Omicron strains and other sarbecoviruses were abrogated after pre-clearing with Wuhan-1 spike protein, antibodies induced by XBB.1.5 boosting in humans focus on conserved epitopes targeted by the antecedent mRNA-1273 primary series. Thus, the antibody response to Omicron-based boosters in humans is imprinted by immunizations with historical mRNA-1273 vaccines, but this outcome may be beneficial as it drives expansion of cross-neutralizing antibodies that inhibit infection of emerging SARS-CoV-2 variants and distantly related sarbecoviruses.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures


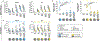
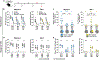


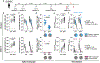


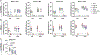
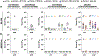



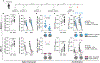

References
MAIN TEXT REFERENCES
METHODS REFERENCES
-
- Buchholz UJ, Finke S & Conzelmann KK Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. Journal of Virology 73, 251–259 (1999). 10.1128/jvi.73.1.251-259.1999 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

