Development and interpretation of a pathomics-driven ensemble model for predicting the response to immunotherapy in gastric cancer
- PMID: 38749538
- PMCID: PMC11097892
- DOI: 10.1136/jitc-2024-008927
Development and interpretation of a pathomics-driven ensemble model for predicting the response to immunotherapy in gastric cancer
Abstract
Background: Only a subset of patients with gastric cancer experience long-term benefits from immune checkpoint inhibitors (ICIs). Currently, there is a deficiency in precise predictive biomarkers for ICI efficacy. The aim of this study was to develop and validate a pathomics-driven ensemble model for predicting the response to ICIs in gastric cancer, using H&E-stained whole slide images (WSI).
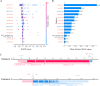
Methods: This multicenter study retrospectively collected and analyzed H&E-stained WSIs and clinical data from 584 patients with gastric cancer. An ensemble model, integrating four classifiers: least absolute shrinkage and selection operator, k-nearest neighbors, decision trees, and random forests, was developed and validated using pathomics features, with the objective of predicting the therapeutic efficacy of immune checkpoint inhibition. Model performance was evaluated using metrics including the area under the curve (AUC), sensitivity, and specificity. Additionally, SHAP (SHapley Additive exPlanations) analysis was used to explain the model's predicted values as the sum of the attribution values for each input feature. Pathogenomics analysis was employed to explain the molecular mechanisms underlying the model's predictions.
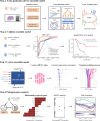
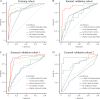
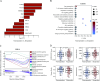
Results: Our pathomics-driven ensemble model effectively stratified the response to ICIs in training cohort (AUC 0.985 (95% CI 0.971 to 0.999)), which was further validated in internal validation cohort (AUC 0.921 (95% CI 0.839 to 0.999)), as well as in external validation cohort 1 (AUC 0.914 (95% CI 0.837 to 0.990)), and external validation cohort 2 (0.927 (95% CI 0.802 to 0.999)). The univariate Cox regression analysis revealed that the prediction signature of pathomics-driven ensemble model was a prognostic factor for progression-free survival in patients with gastric cancer who underwent immunotherapy (p<0.001, HR 0.35 (95% CI 0.24 to 0.50)), and remained an independent predictor after multivariable Cox regression adjusted for clinicopathological variables, (including sex, age, carcinoembryonic antigen, carbohydrate antigen 19-9, therapy regime, line of therapy, differentiation, location and programmed death ligand 1 (PD-L1) expression in all patients (p<0.001, HR 0.34 (95% CI 0.24 to 0.50)). Pathogenomics analysis suggested that the ensemble model is driven by molecular-level immune, cancer, metabolism-related pathways, and was correlated with the immune-related characteristics, including immune score, Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data score, and tumor purity.
Conclusions: Our pathomics-driven ensemble model exhibited high accuracy and robustness in predicting the response to ICIs using WSIs. Therefore, it could serve as a novel and valuable tool to facilitate precision immunotherapy.
Keywords: Gastric Cancer; Immune Checkpoint Inhibitor; Pathology.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
-
- Shitara K, Van Cutsem E, Bang Y-J, et al. . Efficacy and safety of pembrolizumab or pembrolizumab plus chemotherapy vs chemotherapy alone for patients with first-line, advanced gastric cancer: the KEYNOTE-062 phase 3 randomized clinical trial. JAMA Oncol 2020;6:1571–80. 10.1001/jamaoncol.2020.3370 - DOI - PMC - PubMed
-
- Janjigian YY, Shitara K, Moehler M, et al. . First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (Checkmate 649): a randomised, open-label, phase 3 trial. Lancet 2021;398:27–40. 10.1016/S0140-6736(21)00797-2 - DOI - PMC - PubMed
-
- Salas-Benito D, Pérez-Gracia JL, Ponz-Sarvisé M, et al. . Paradigms on immunotherapy combinations with chemotherapy. Cancer Discov 2021;11:1353–67. 10.1158/2159-8290.CD-20-1312 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
