Octyl itaconate enhances VSVΔ51 oncolytic virotherapy by multitarget inhibition of antiviral and inflammatory pathways
- PMID: 38750019
- PMCID: PMC11096414
- DOI: 10.1038/s41467-024-48422-x
Octyl itaconate enhances VSVΔ51 oncolytic virotherapy by multitarget inhibition of antiviral and inflammatory pathways
Erratum in
-
Author Correction: Octyl itaconate enhances VSVΔ51 oncolytic virotherapy by multitarget inhibition of antiviral and inflammatory pathways.Nat Commun. 2024 Dec 19;15(1):10698. doi: 10.1038/s41467-024-55044-w. Nat Commun. 2024. PMID: 39702301 Free PMC article. No abstract available.
Abstract
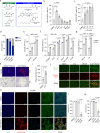
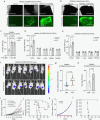
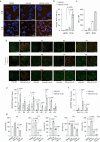
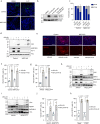
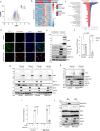
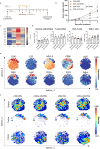
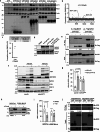
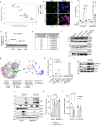
The presence of heterogeneity in responses to oncolytic virotherapy poses a barrier to clinical effectiveness, as resistance to this treatment can occur through the inhibition of viral spread within the tumor, potentially leading to treatment failures. Here we show that 4-octyl itaconate (4-OI), a chemical derivative of the Krebs cycle-derived metabolite itaconate, enhances oncolytic virotherapy with VSVΔ51 in various models including human and murine resistant cancer cell lines, three-dimensional (3D) patient-derived colon tumoroids and organotypic brain tumor slices. Furthermore, 4-OI in combination with VSVΔ51 improves therapeutic outcomes in a resistant murine colon tumor model. Mechanistically, we find that 4-OI suppresses antiviral immunity in cancer cells through the modification of cysteine residues in MAVS and IKKβ independently of the NRF2/KEAP1 axis. We propose that the combination of a metabolite-derived drug with an oncolytic virus agent can greatly improve anticancer therapeutic outcomes by direct interference with the type I IFN and NF-κB-mediated antiviral responses.
© 2024. The Author(s).
Conflict of interest statement
M.M.R. reports research funding from Novo Nordisk A/S. The remaining authors declare no competing interests.
Figures
References
-
- Pol, J. G., Workenhe, S. T., Konda, P., Gujar, S. & Kroemer, G. Cytokines in oncolytic virotherapy. Cytokine Growth Factor Rev.56, 4–27 (2020). - PubMed
-
- Twumasi-Boateng, K., Pettigrew, J. L., Kwok, Y. Y. E., Bell, J. C. & Nelson, B. H. Oncolytic viruses as engineering platforms for combination immunotherapy. Nat. Rev. Cancer18, 419–432 (2018). - PubMed
MeSH terms
Grants and funding
- 11056/KWF Kankerbestrijding (Dutch Cancer Society)
- R335-2019-2138/Lundbeckfonden (Lundbeck Foundation)
- R325-2019-1490/Lundbeckfonden (Lundbeck Foundation)
- NNF22OC0079512/Novo Nordisk Fonden (Novo Nordisk Foundation)
- R279-A16218/Kræftens Bekæmpelse (Danish Cancer Society)
- IG-2019-22891/Associazione Italiana di Oncologia Medica (Italian Association of Medical Oncology)
- DNRF164/Danmarks Grundforskningsfond (Danish National Research Foundation)
- III L 5 - 519/03/03.001 - (0015)/Hessisches Ministerium für Wissenschaft und Kunst (Hessen State Ministry of Higher Education, Research and the Arts)
- 22869/CRUK_/Cancer Research UK/United Kingdom
- NNF19OC0054782/Novo Nordisk Fonden (Novo Nordisk Foundation)
- R307-2018-3667/Lundbeckfonden (Lundbeck Foundation)
- DFF-37741/Det Frie Forskningsråd (Danish Council for Independent Research)
- CF19-0687/Carlsbergfondet (Carlsberg Foundation)
- C52419/A22869/Cancer Research UK (CRUK)
- NNF19OC0056043/Novo Nordisk Fonden (Novo Nordisk Foundation)
- 1026-00003B/Det Frie Forskningsråd (Danish Council for Independent Research)
- R295-A16770/Dansk Kræftforsknings Fond (Danish Cancer Research Fund)
- R306-A18092/Kræftens Bekæmpelse (Danish Cancer Society)
- 9039-00307B/Det Frie Forskningsråd (Danish Council for Independent Research)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

