Nuclear actin structure regulates chromatin accessibility
- PMID: 38750021
- PMCID: PMC11096319
- DOI: 10.1038/s41467-024-48580-y
Nuclear actin structure regulates chromatin accessibility
Abstract
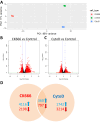
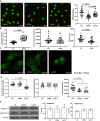
Polymerized β-actin may provide a structural basis for chromatin accessibility and actin transport into the nucleus can guide mesenchymal stem cell (MSC) differentiation. Using MSC, we show that using CK666 to inhibit Arp2/3 directed secondary actin branching results in decreased nuclear actin structure, and significantly alters chromatin access measured with ATACseq at 24 h. The ATAC-seq results due to CK666 are distinct from those caused by cytochalasin D (CytoD), which enhances nuclear actin structure. In addition, nuclear visualization shows Arp2/3 inhibition decreases pericentric H3K9me3 marks. CytoD, alternatively, induces redistribution of H3K27me3 marks centrally. Such alterations in chromatin landscape are consistent with differential gene expression associated with distinctive differentiation patterns. Further, knockdown of the non-enzymatic monomeric actin binding protein, Arp4, leads to extensive chromatin unpacking, but only a modest increase in transcription, indicating an active role for actin-Arp4 in transcription. These data indicate that dynamic actin remodeling can regulate chromatin interactions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Grants and funding
- AR076803/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- AR073264/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- AG069923/U.S. Department of Health & Human Services | NIH | Center for Scientific Review (NIH Center for Scientific Review)
- R01 AR073264/AR/NIAMS NIH HHS/United States
- S10 OD032354/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

