Emerging variants, unique phenotypes, and transcriptomic signatures: an integrated study of COASY-associated diseases
- PMID: 38750253
- PMCID: PMC11187879
- DOI: 10.1002/acn3.52079
Emerging variants, unique phenotypes, and transcriptomic signatures: an integrated study of COASY-associated diseases
Abstract
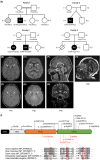
Objective: COASY, the gene encoding the bifunctional enzyme CoA synthase, which catalyzes the last two reactions of cellular de novo coenzyme A (CoA) biosynthesis, has been linked to two exceedingly rare autosomal recessive disorders, such as COASY protein-associated neurodegeneration (CoPAN), a form of neurodegeneration with brain iron accumulation (NBIA), and pontocerebellar hypoplasia type 12 (PCH12). We aimed to expand the phenotypic spectrum and gain insights into the pathogenesis of COASY-related disorders.
Methods: Patients were identified through targeted or exome sequencing. To unravel the molecular mechanisms of disease, RNA sequencing, bioenergetic analysis, and quantification of critical proteins were performed on fibroblasts.
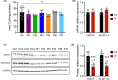
Results: We identified five new individuals harboring novel COASY variants. While one case exhibited classical CoPAN features, the others displayed atypical symptoms such as deafness, language and autism spectrum disorders, brain atrophy, and microcephaly. All patients experienced epilepsy, highlighting its potential frequency in COASY-related disorders. Fibroblast transcriptomic profiling unveiled dysregulated expression in genes associated with mitochondrial respiration, responses to oxidative stress, transmembrane transport, various cellular signaling pathways, and protein translation, modification, and trafficking. Bioenergetic analysis revealed impaired mitochondrial oxygen consumption in COASY fibroblasts. Despite comparable total CoA levels to control cells, the amounts of mitochondrial 4'-phosphopantetheinylated proteins were significantly reduced in COASY patients.
Interpretation: These results not only extend the clinical phenotype associated with COASY variants but also suggest a continuum between CoPAN and PCH12. The intricate interplay of altered cellular processes and signaling pathways provides valuable insights for further research into the pathogenesis of COASY-associated diseases.
© 2024 The Authors. Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Zhyvoloup A, Nemazanyy I, Babich A, et al. Molecular cloning of CoA synthase. The missing link in CoA biosynthesis. J Biol Chem. 2002;277(25):22107‐22110. - PubMed
-
- Zhyvoloup A, Nemazanyy I, Panasyuk G, et al. Subcellular localization and regulation of coenzyme A synthase. J Biol Chem. 2003;278(50):50316‐50321. - PubMed
-
- Nemazanyy I, Panasyuk G, Breus O, Zhyvoloup A, Filonenko V, Gout IT. Identification of a novel CoA synthase isoform, which is primarily expressed in the brain. Biochem Biophys Res Commun. 2006;341(4):995‐1000. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

