From pixels to connections: exploring in vitro neuron reconstruction software for network graph generation
- PMID: 38750282
- PMCID: PMC11096190
- DOI: 10.1038/s42003-024-06264-9
From pixels to connections: exploring in vitro neuron reconstruction software for network graph generation
Abstract
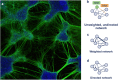
Digital reconstruction has been instrumental in deciphering how in vitro neuron architecture shapes information flow. Emerging approaches reconstruct neural systems as networks with the aim of understanding their organization through graph theory. Computational tools dedicated to this objective build models of nodes and edges based on key cellular features such as somata, axons, and dendrites. Fully automatic implementations of these tools are readily available, but they may also be purpose-built from specialized algorithms in the form of multi-step pipelines. Here we review software tools informing the construction of network models, spanning from noise reduction and segmentation to full network reconstruction. The scope and core specifications of each tool are explicitly defined to assist bench scientists in selecting the most suitable option for their microscopy dataset. Existing tools provide a foundation for complete network reconstruction, however more progress is needed in establishing morphological bases for directed/weighted connectivity and in software validation.
© 2024. Crown.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

