Combining germline, tissue and liquid biopsy analysis by comprehensive genomic profiling to improve the yield of actionable variants in a real-world cancer cohort
- PMID: 38750555
- PMCID: PMC11097509
- DOI: 10.1186/s12967-024-05227-2
Combining germline, tissue and liquid biopsy analysis by comprehensive genomic profiling to improve the yield of actionable variants in a real-world cancer cohort
Abstract
Background: Comprehensive next-generation sequencing is widely used for precision oncology and precision prevention approaches. We aimed to determine the yield of actionable gene variants, the capacity to uncover hereditary predisposition and liquid biopsy appropriateness instead of, or in addition to, tumor tissue analysis, in a real-world cohort of cancer patients, who may benefit the most from comprehensive genomic profiling.
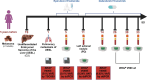
Methods: Seventy-eight matched germline/tumor tissue/liquid biopsy DNA and RNA samples were profiled using the Hereditary Cancer Panel (germline) and the TruSight Oncology 500 panel (tumor tissue/cfDNA) from 23 patients consecutively enrolled at our center according to at least one of the following criteria: no available therapeutic options; long responding patients potentially fit for other therapies; rare tumor; suspected hereditary cancer; primary cancer with high metastatic potential; tumor of unknown primary origin. Variants were annotated for OncoKB and AMP/ASCO/CAP classification.
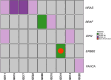
Results: The overall yield of actionable somatic and germline variants was 57% (13/23 patients), and 43.5%, excluding variants previously identified by somatic or germline routine testing. The accuracy of tumor/cfDNA germline-focused analysis was demonstrated by overlapping results of germline testing. Five germline variants in BRCA1, VHL, CHEK1, ATM genes would have been missed without extended genomic profiling. A previously undetected BRAF p.V600E mutation was emblematic of the clinical utility of this approach in a patient with a liver undifferentiated embryonal sarcoma responsive to BRAF/MEK inhibition.
Conclusions: Our study confirms the clinical relevance of performing extended parallel tumor DNA and cfDNA testing to broaden therapeutic options, to longitudinally monitor cfDNA during patient treatment, and to uncover possible hereditary predisposition following tumor sequencing in patient care.
Keywords: Circulating cell-free DNA; Clinically actionable variants; Droplet digital pcr (ddPCR); Germline pathogenic variants; Next-generation sequencing; Targeted therapy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no financial or non‐financial competing interest.
Figures


References
-
- Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(11):1491–1505. doi: 10.1016/j.annonc.2020.07.014. - DOI - PubMed
-
- Hoes LR, Van Berge Henegouwen JM, Van Der Wijngaart H, Zeverijn LJ, Van Der Velden DL, Van De Haar J, et al. Patients with rare cancers in the drug rediscovery protocol (DRUP) benefit from genomics-guided treatment. Clin Cancer Res. 2022;28(7):1402–11. doi: 10.1158/1078-0432.CCR-21-3752. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- IRCCS Ospedale Policlinico San Martino 5×1000 5M-2019-23680284/Ministero della Salute
- IRCCS Ospedale Policlinico San Martino 5×1000 5M-2020-23682092/Ministero della Salute
- IRCCS Ospedale Policlinico San Martino Ricerca Corrente 2022 n.2772539/Ministero della Salute
- IRCCS Ospedale Policlinico San Martino Ricerca Corrente n.72562/Ministero della Salute
- Ricerca Corrente 2023/Ministero della Salute
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

