NF-κB Inducing Kinase Attenuates Colorectal Cancer by Regulating Noncanonical NF-κB Mediated Colonic Epithelial Cell Regeneration
- PMID: 38750899
- PMCID: PMC11278896
- DOI: 10.1016/j.jcmgh.2024.05.004
NF-κB Inducing Kinase Attenuates Colorectal Cancer by Regulating Noncanonical NF-κB Mediated Colonic Epithelial Cell Regeneration
Abstract
Background & aims: Dysregulated colonic epithelial cell (CEC) proliferation is a critical feature in the development of colorectal cancer. We show that NF-κB-inducing kinase (NIK) attenuates colorectal cancer through coordinating CEC regeneration/differentiation via noncanonical NF-κB signaling that is unique from canonical NF-kB signaling.
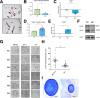
Methods: Initial studies evaluated crypt morphology/functionality, organoid generation, transcriptome profiles, and the microbiome. Inflammation and inflammation-induced tumorigenesis were initiated in whole-body NIK knockout mice (Nik-/-) and conditional-knockout mice following administration of azoxymethane and dextran sulfate sodium.
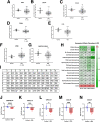
Results: Human transcriptomic data revealed dysregulated noncanonical NF-kB signaling. In vitro studies evaluating Nik-/- crypts and organoids derived from mature, nondividing CECs, and colonic stem cells exhibited increased accumulation and stunted growth, respectively. Transcriptomic analysis of Nik-/- cells revealed gene expression signatures associated with altered differentiation-regeneration. When assessed in vivo, Nik-/- mice exhibited more severe colitis with dextran sulfate sodium administration and an altered microbiome characterized by increased colitogenic microbiota. In the inflammation-induced tumorigenesis model, we observed both increased tumor burdens and inflammation in mice where NIK is knocked out in CECs (NikΔCEC). Interestingly, this was not recapitulated when NIK was conditionally knocked out in myeloid cells (NikΔMYE). Surprisingly, conditional knockout of the canonical pathway in myeloid cells (RelAΔMYE) revealed decreased tumor burden and inflammation and no significant changes when conditionally knocked out in CECs (RelAΔCEC).
Conclusions: Dysregulated noncanonical NF-κB signaling is associated with the development of colorectal cancer in a tissue-dependent manner and defines a critical role for NIK in regulating gastrointestinal inflammation and regeneration associated with colorectal cancer.
Keywords: Colitis-Associated Tumorigenesis; Differentiation; Epithelial Cells; Organoids.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

