Comprehensive analysis of juvenile idiopathic arthritis patients' immune characteristics based on bulk and single-cell sequencing data
- PMID: 38751447
- PMCID: PMC11094213
- DOI: 10.3389/fmolb.2024.1359235
Comprehensive analysis of juvenile idiopathic arthritis patients' immune characteristics based on bulk and single-cell sequencing data
Abstract
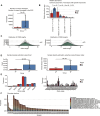
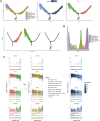
Background: The pathogenesis of juvenile idiopathic arthritis (JIA) is strongly influenced by an impaired immune system. However, the molecular mechanisms underlying its development and progression have not been elucidated. In this study, the computational methods TRUST4 were used to construct a T-cell receptor (TCR) and B-cell receptor (BCR) repertoire from the peripheral blood of JIA patients via bulk RNA-seq data, after which the clonality and diversity of the immune repertoire were analyzed.
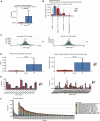
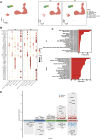
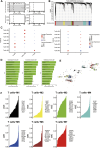
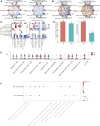
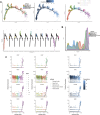
Results: Our findings revealed significant differences in the frequency of clonotypes between the JIA and healthy control groups in terms of the TCR and BCR repertoires. This work identified specific V genes and J genes in TCRs and BCRs that could be used to expand our understanding of JIA. After single-cell RNA analysis, the relative percentages of CD14 monocytes were significantly greater in the JIA group. Cell-cell communication analysis revealed the significant role of the MIF signaling pathway in JIA.
Conclusion: In conclusion, this work describes the immune features of both the TCR and BCR repertoires under JIA conditions and provides novel insight into immunotherapy for JIA.
Keywords: B-cell receptor; T-cell receptor; immune repertoire; juvenile idiopathic arthritis; single-cell RNA sequencing.
Copyright © 2024 Liu, Gong, Lin and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Revealing the Immune Heterogeneity between Systemic Lupus Erythematosus and Rheumatoid Arthritis Based on Multi-Omics Data Analysis.Int J Mol Sci. 2022 May 5;23(9):5166. doi: 10.3390/ijms23095166. Int J Mol Sci. 2022. PMID: 35563556 Free PMC article.
-
Large-scale integrative analysis of juvenile idiopathic arthritis for new insight into its pathogenesis.Arthritis Res Ther. 2024 Feb 10;26(1):47. doi: 10.1186/s13075-024-03280-2. Arthritis Res Ther. 2024. PMID: 38336809 Free PMC article.
-
Next-Generation Sequencing Reveals Restriction and Clonotypic Expansion of Treg Cells in Juvenile Idiopathic Arthritis.Arthritis Rheumatol. 2016 Jul;68(7):1758-68. doi: 10.1002/art.39606. Arthritis Rheumatol. 2016. PMID: 26815131 Free PMC article.
-
Comprehensive Analysis of TCR and BCR Repertoires: Insights into Methodologies, Challenges, and Applications.Genomics Inform. 2025 Feb 24;23(1):6. doi: 10.1186/s44342-024-00034-z. Genomics Inform. 2025. PMID: 39994831 Free PMC article. Review.
-
Single-cell RNA sequencing in juvenile idiopathic arthritis.Genes Dis. 2023 May 24;11(2):633-644. doi: 10.1016/j.gendis.2023.04.014. eCollection 2024 Mar. Genes Dis. 2023. PMID: 37692495 Free PMC article. Review.
Cited by
-
Comprehensive Characterization of Th2/Th17 Cells-Related Gene in Systemic Juvenile Rheumatoid Arthritis: Evidence from Mendelian Randomization and Transcriptome Data Using Multiple Machine Learning Approaches.Int J Gen Med. 2024 Dec 10;17:5973-5996. doi: 10.2147/IJGM.S482288. eCollection 2024. Int J Gen Med. 2024. PMID: 39678686 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

