Innovative Reversed-Phase Chromatography Platform Approach for the Fast and Accurate Characterization of Membrane Vesicles' Protein Patterns
- PMID: 38751636
- PMCID: PMC11091982
- DOI: 10.1021/acsptsci.4c00112
Innovative Reversed-Phase Chromatography Platform Approach for the Fast and Accurate Characterization of Membrane Vesicles' Protein Patterns
Abstract
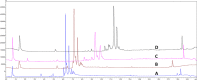
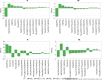
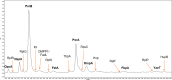
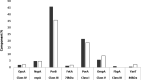
Outer membrane vesicles (OMVs) have been widely explored to develop vaccine candidates for bacterial pathogens due to their ability to combine adjuvant properties with immunogenic activity. OMV expresses a variety of proteins and carbohydrate antigens on their surfaces. For this reason, there is an analytical need to thoroughly characterize the species expressed at their surface: we here present a simple and accurate reversed-phase ultrahigh-performance liquid chromatography (RP-UPLC) method developed according to quality by design principles. This work provides an analytical alternative to the classical sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) characterization. The higher selectivity and sensitivity of the RP-UHPLC assay allow for the identification of additional protein species with respect to SDS-PAGE and facilitate its precise relative abundance quantification. According to validation results, the assay showed high accuracy, linearity, precision, repeatability, and a limit of quantification of 1% for less abundant proteins. This performance paves the way for improved production campaign consistency while also being analytically simple (no sample pretreatment required), making it suitable for routine quality control testing. In addition, the applicability of the assay to a wider range of vesicle classes (GMMA) was demonstrated.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
LinkOut - more resources
Full Text Sources
