Screening and identification of an aflatoxin B1-degrading strain from the Qinghai-Tibet Plateau and biodegradation products analysis
- PMID: 38751722
- PMCID: PMC11094616
- DOI: 10.3389/fmicb.2024.1367297
Screening and identification of an aflatoxin B1-degrading strain from the Qinghai-Tibet Plateau and biodegradation products analysis
Abstract
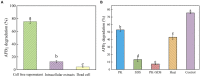
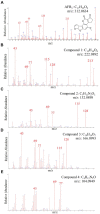
This research aimed to address the issue of aflatoxin B1 (AFB1) contamination, which posed severe health and economic consequences. This study involved exploring unique species resources in the Qinghai-Tibet Plateau, screening strains capable of degrading AFB1. UPLC-Q-Orbitrap HRMS and NMR were employed to examine the degradation process and identify the structure of the degradation products. Results showed that Bacillus amyloliquefaciens YUAD7, isolated from yak dung in the Qinghai-Tibet Plateau, removed 91.7% of AFB1 from TSB-AFB1 medium with an AFB1 concentration of 10 μg/mL (72 h, 37°C, pH 6.8) and over 85% of AFB1 from real food samples at 10 μg/g (72 h, 37°C), exhibiting strong AFB1 degradation activity. Bacillus amyloliquefaciens YUAD7's extracellular secretions played a major role in AFB1 degradation mediated and could still degrade AFB1 by 43.16% after boiling for 20 min. Moreover, B. amyloliquefaciens YUAD7 demonstrated the capability to decompose AFB1 through processes such as hydrogenation, enzyme modification, and the elimination of the -CO group, resulting in the formation of smaller non-toxic molecules. Identified products include C12H14O4, C5H12N2O2, C10H14O2, C4H12N2O, with a structure consisting of dimethoxyphenyl and enoic acid, dimethyl-amino and ethyl carbamate, polyunsaturated fatty acid, and aminomethyl. The results indicated that B. amyloliquefaciens YUAD7 could be a potentially valuable strain for industrial-scale biodegradation of AFB1 and providing technical support and new perspectives for research on biodegradation products.
Keywords: Bacillus amyloliquefaciens YUAD7; aflatoxin B1; biological detoxification; degradation products; detoxification mechanism.
Copyright © 2024 Tang, Liu, Dong and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Detoxification of Aflatoxin B1 by a Potential Probiotic Bacillus amyloliquefaciens WF2020.Front Microbiol. 2022 May 10;13:891091. doi: 10.3389/fmicb.2022.891091. eCollection 2022. Front Microbiol. 2022. PMID: 35620100 Free PMC article.
-
Biodegradation Characteristics and Mechanism of Aflatoxin B1 by Bacillus amyloliquefaciens from Enzymatic and Multiomics Perspectives.J Agric Food Chem. 2024 Jul 17;72(28):15841-15853. doi: 10.1021/acs.jafc.4c04055. Epub 2024 Jul 3. J Agric Food Chem. 2024. PMID: 38957116
-
Isolation, Purification, and Characterization of a Laccase-Degrading Aflatoxin B1 from Bacillus amyloliquefaciens B10.Toxins (Basel). 2022 Mar 31;14(4):250. doi: 10.3390/toxins14040250. Toxins (Basel). 2022. PMID: 35448859 Free PMC article.
-
Identification of a Novel Aflatoxin B1-Degrading Strain, Bacillus halotolerans DDC-4, and Its Response Mechanisms to Aflatoxin B1.Toxins (Basel). 2024 May 31;16(6):256. doi: 10.3390/toxins16060256. Toxins (Basel). 2024. PMID: 38922150 Free PMC article.
-
Mechanisms and transformed products of aflatoxin B1 degradation under multiple treatments: a review.Crit Rev Food Sci Nutr. 2024;64(8):2263-2275. doi: 10.1080/10408398.2022.2121910. Epub 2022 Sep 14. Crit Rev Food Sci Nutr. 2024. PMID: 36102160 Review.
Cited by
-
Mycotoxin Biodegradation by Bacillus Bacteria-A Review.Toxins (Basel). 2024 Nov 4;16(11):478. doi: 10.3390/toxins16110478. Toxins (Basel). 2024. PMID: 39591233 Free PMC article. Review.
-
Indole-3-acetic acid enhances ruminal microbiota for aflatoxin B1 removal in vitro fermentation.Front Vet Sci. 2024 Dec 20;11:1450241. doi: 10.3389/fvets.2024.1450241. eCollection 2024. Front Vet Sci. 2024. PMID: 39758608 Free PMC article.
-
Characterization and genomic analysis of Bacillus megaterium with the ability to degrade aflatoxin B1.Front Microbiol. 2024 Aug 7;15:1407270. doi: 10.3389/fmicb.2024.1407270. eCollection 2024. Front Microbiol. 2024. PMID: 39171271 Free PMC article.
References
-
- Adebo O. A., Njobeh P. B., Mavumengwana V. (2016). Degradation and detoxification of AFB1 by Staphylocococcus warneri, Sporosarcina sp. and Lysinibacillus fusiformis. Food Control 68, 92–96. doi: 10.1016/j.foodcont.2016.03.021 - DOI
-
- Bai J., Ding Z., Ke W., Xu D., Wang M., Huang W., et al. . (2021). Different lactic acid bacteria and their combinations regulated the fermentation process of ensiled alfalfa: ensiling characteristics, dynamics of bacterial community and their functional shifts. J. Microbial. Biotechnol. 14, 1171–1182. doi: 10.1111/1751-7915.13785, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

