Two extracellular α-arabinofuranosidases are required for cereal-derived arabinoxylan metabolism by Bifidobacterium longum subsp. longum
- PMID: 38752423
- PMCID: PMC11318964
- DOI: 10.1080/19490976.2024.2353229
Two extracellular α-arabinofuranosidases are required for cereal-derived arabinoxylan metabolism by Bifidobacterium longum subsp. longum
Abstract
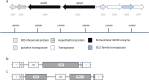
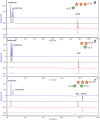
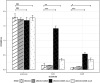
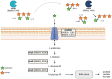
Members of the genus Bifidobacterium are commonly found in the human gut and are known to utilize complex carbohydrates that are indigestible by the human host. Members of the Bifidobacterium longum subsp. longum taxon can metabolize various plant-derived carbohydrates common to the human diet. To metabolize such polysaccharides, which include arabinoxylan, bifidobacteria need to encode appropriate carbohydrate-active enzymes in their genome. In the current study, we describe two GH43 family enzymes, denoted here as AxuA and AxuB, which are encoded by B. longum subsp. longum NCIMB 8809 and are shown to be required for cereal-derived arabinoxylan metabolism by this strain. Based on the observed hydrolytic activity of AxuA and AxuB, assessed by employing various synthetic and natural substrates, and based on in silico analyses, it is proposed that both AxuA and AxuB represent extracellular α-L-arabinofuranosidases with distinct substrate preferences. The variable presence of the axuA and axuB genes and other genes previously described to be involved in the metabolism of arabinose-containing glycans can in the majority cases explain the (in)ability of individual B. longum subsp. longum strains to grow on cereal-derived arabinoxylans and arabinan.
Keywords: Dietary fiber; bifidobacterial; gut microbiota; prebiotic; probiotic.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures





Similar articles
-
Two Novel α-l-Arabinofuranosidases from Bifidobacterium longum subsp. longum Belonging to Glycoside Hydrolase Family 43 Cooperatively Degrade Arabinan.Appl Environ Microbiol. 2019 Mar 6;85(6):e02582-18. doi: 10.1128/AEM.02582-18. Print 2019 Mar 15. Appl Environ Microbiol. 2019. PMID: 30635377 Free PMC article.
-
Multiple Transporters and Glycoside Hydrolases Are Involved in Arabinoxylan-Derived Oligosaccharide Utilization in Bifidobacterium pseudocatenulatum.Appl Environ Microbiol. 2020 Nov 24;86(24):e01782-20. doi: 10.1128/AEM.01782-20. Print 2020 Nov 24. Appl Environ Microbiol. 2020. PMID: 33036985 Free PMC article.
-
Two α-L-arabinofuranosidases from Bifidobacterium longum subsp. longum are involved in arabinoxylan utilization.Appl Microbiol Biotechnol. 2022 Mar;106(5-6):1957-1965. doi: 10.1007/s00253-022-11845-x. Epub 2022 Mar 2. Appl Microbiol Biotechnol. 2022. PMID: 35235007
-
Bifidobacterium carbohydrases-their role in breakdown and synthesis of (potential) prebiotics.Mol Nutr Food Res. 2008 Jan;52(1):146-63. doi: 10.1002/mnfr.200700121. Mol Nutr Food Res. 2008. PMID: 18040988 Review.
-
Plant Glycan Metabolism by Bifidobacteria.Front Microbiol. 2021 Feb 4;12:609418. doi: 10.3389/fmicb.2021.609418. eCollection 2021. Front Microbiol. 2021. PMID: 33613480 Free PMC article. Review.
Cited by
-
Integrative genomic reconstruction reveals heterogeneity in carbohydrate utilization across human gut bifidobacteria.bioRxiv [Preprint]. 2025 May 17:2024.07.06.602360. doi: 10.1101/2024.07.06.602360. bioRxiv. 2025. Update in: Nat Microbiol. 2025 Aug;10(8):2031-2047. doi: 10.1038/s41564-025-02056-x. PMID: 39005317 Free PMC article. Updated. Preprint.
-
Gene-trait matching among Bifidobacterium dentium strains reveals various glycan metabolism loci including a strain-specific fucosyllactose utilization cluster.Front Microbiol. 2025 May 12;16:1584694. doi: 10.3389/fmicb.2025.1584694. eCollection 2025. Front Microbiol. 2025. PMID: 40421466 Free PMC article.
-
Integrative genomic reconstruction reveals heterogeneity in carbohydrate utilization across human gut bifidobacteria.Nat Microbiol. 2025 Aug;10(8):2031-2047. doi: 10.1038/s41564-025-02056-x. Epub 2025 Jul 16. Nat Microbiol. 2025. PMID: 40670725 Free PMC article.
References
-
- Arboleya S, Bottacini F, O’Connell-Motherway M, Ryan CA, Ross RP, van Sinderen D, Stanton C. Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genomics. 2018;19(1). doi: 10.1186/s12864-017-4388-9. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
