Two residues in the DNA binding site of Pif1 helicase are essential for nuclear functions but dispensable for mitochondrial respiratory growth
- PMID: 38752483
- PMCID: PMC11194084
- DOI: 10.1093/nar/gkae403
Two residues in the DNA binding site of Pif1 helicase are essential for nuclear functions but dispensable for mitochondrial respiratory growth
Abstract
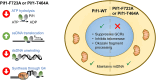
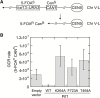
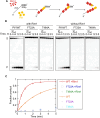
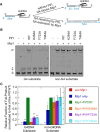
Pif1 helicase functions in both the nucleus and mitochondria. Pif1 tightly couples ATP hydrolysis, single-stranded DNA translocation, and duplex DNA unwinding. We investigated two Pif1 variants (F723A and T464A) that have each lost one site of interaction of the protein with the DNA substrate. Both variants exhibit minor reductions in affinity for DNA and ATP hydrolysis but have impaired DNA unwinding activity. However, these variants translocate on single-stranded DNA faster than the wildtype enzyme and can slide on the DNA substrate in an ATP-independent manner. This suggests they have lost their grip on the DNA, interfering with coupling ATP hydrolysis to translocation and unwinding. Yeast expressing these variants have increased gross chromosomal rearrangements, increased telomere length, and can overcome the lethality of dna2Δ, similar to phenotypes of yeast lacking Pif1. However, unlike pif1Δ mutants, they are viable on glycerol containing media and maintain similar mitochondrial DNA copy numbers as Pif1 wildtype. Overall, our data indicate that a tight grip of the trailing edge of the Pif1 enzyme on the DNA couples ATP hydrolysis to DNA translocation and DNA unwinding. This tight grip appears to be essential for the Pif1 nuclear functions tested but is dispensable for mitochondrial respiratory growth.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

