In Silico Prediction of Quercetin Analogs for Targeting Death-Associated Protein Kinase 1 (DAPK1) Against Alzheimer's Disease
- PMID: 38752632
- PMCID: PMC11451310
- DOI: 10.2174/1570159X22666240515090434
In Silico Prediction of Quercetin Analogs for Targeting Death-Associated Protein Kinase 1 (DAPK1) Against Alzheimer's Disease
Abstract
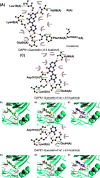
Alzheimer's Disease (AD) is a progressive neurodegenerative disorder that greatly affects the health and life quality of the elderly population. Existing drugs mainly alleviate symptoms but fail to halt disease progression, underscoring the urgent need for the development of novel drugs. Based on the neuroprotective effects of flavonoid quercetin in AD, this study was designed to identify potential AD-related targets for quercetin and perform in silico prediction of promising analogs for the treatment of AD. Database mining suggested death-associated protein kinase 1 (DAPK1) as the most promising AD-related target for quercetin among seven protein candidates. To achieve better biological effects for the treatment of AD, we devised a series of quercetin analogs as ligands for DAPK1, and molecular docking analyses, absorption, distribution, metabolism, and excretion (ADME) predictions, as well as molecular dynamics (MD) simulations, were performed. The energy for drug-protein interaction was predicted and ranked. As a result, quercetin-A1a and quercetin-A1a1 out of 19 quercetin analogs exhibited the lowest interaction energy for binding to DAPK1 than quercetin, and they had similar dynamics performance with quercetin. In addition, quercetin-A1a and quercetin-A1a1 were predicted to have better water solubility. Thus, quercetin-A1a and quercetin-A1a1 could be promising agents for the treatment of AD. Our findings paved the way for further experimental studies and the development of novel drugs.
Keywords: Alzheimer’s disease (AD); In silico prediction; death-associated protein kinase 1 (DAPK1); neurodegenerative disease.; quercetin; quercetin analogs.
Conflict of interest statement
The authors declare no conflict of interest, financial or otherwise.
Figures



References
-
- Nichols E., Szoeke C.E.I., Vollset S.E., Abbasi N., Abd-Allah F., Abdela J., Aichour M.T.E., Akinyemi R.O., Alahdab F., Asgedom S.W., Awasthi A., Barker-Collo S.L., Baune B.T., Béjot Y., Belachew A.B., Bennett D.A., Biadgo B., Bijani A., Bin Sayeed M.S., Brayne C., Carpenter D.O., Carvalho F., Catalá-López F., Cerin E., Choi J-Y.J., Dang A.K., Degefa M.G., Djalalinia S., Dubey M., Duken E.E., Edvardsson D., Endres M., Eskandarieh S., Faro A., Farzadfar F., Fereshtehnejad S-M., Fernandes E., Filip I., Fischer F., Gebre A.K., Geremew D., Ghasemi-Kasman M., Gnedovskaya E.V., Gupta R., Hachinski V., Hagos T.B., Hamidi S., Hankey G.J., Haro J.M., Hay S.I., Irvani S.S.N., Jha R.P., Jonas J.B., Kalani R., Karch A., Kasaeian A., Khader Y.S., Khalil I.A., Khan E.A., Khanna T., Khoja T.A.M., Khubchandani J., Kisa A., Kissimova-Skarbek K., Kivimäki M., Koyanagi A., Krohn K.J., Logroscino G., Lorkowski S., Majdan M., Malekzadeh R., März W., Massano J., Mengistu G., Meretoja A., Mohammadi M., Mohammadi-Khanaposhtani M., Mokdad A.H., Mondello S., Moradi G., Nagel G., Naghavi M., Naik G., Nguyen L.H., Nguyen T.H., Nirayo Y.L., Nixon M.R., Ofori-Asenso R., Ogbo F.A., Olagunju A.T., Owolabi M.O., Panda-Jonas S., Passos V.M.A., Pereira D.M., Pinilla-Monsalve G.D., Piradov M.A., Pond C.D., Poustchi H., Qorbani M., Radfar A., Reiner R.C., Jr, Robinson S.R., Roshandel G., Rostami A., Russ T.C., Sachdev P.S., Safari H., Safiri S., Sahathevan R., Salimi Y., Satpathy M., Sawhney M., Saylan M., Sepanlou S.G., Shafieesabet A., Shaikh M.A., Sahraian M.A., Shigematsu M., Shiri R., Shiue I., Silva J.P., Smith M., Sobhani S., Stein D.J., Tabarés-Seisdedos R., Tovani-Palone M.R., Tran B.X., Tran T.T., Tsegay A.T., Ullah I., Venketasubramanian N., Vlassov V., Wang Y-P., Weiss J., Westerman R., Wijeratne T., Wyper G.M.A., Yano Y., Yimer E.M., Yonemoto N., Yousefifard M., Zaidi Z., Zare Z., Vos T., Feigin V.L., Murray C.J.L. Global, regional, and national burden of Alzheimer’s disease and other dementias, 1990-2016: A systematic analysis for the global burden of disease study 2016. Lancet Neurol. 2019;18(1):88–106. doi: 10.1016/S1474-4422(18)30403-4. - DOI - PMC - PubMed
-
- 2020 Alzheimer’s disease facts and figures. Alzheimers Dement. 2020:2020. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

