IL-24 promotes atopic dermatitis-like inflammation through driving MRSA-induced allergic responses
- PMID: 38752989
- PMCID: PMC11892005
- DOI: 10.1093/procel/pwae030
IL-24 promotes atopic dermatitis-like inflammation through driving MRSA-induced allergic responses
Abstract
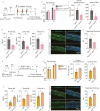
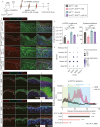
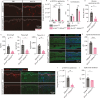
Atopic dermatitis (AD) is a prevalent inflammatory skin disorder in which patients experience recurrent eczematous lesions and intense itching. The colonization of Staphylococcus aureus (S. aureus) is correlated with the severity of the disease, but its role in AD development remains elusive. Using single-cell RNA sequencing, we uncovered that keratinocytes activate a distinct immune response characterized by induction of Il24 when exposed to methicillin-resistant S. aureus (MRSA). Further experiments using animal models showed that the administration of recombinant IL-24 protein worsened AD-like pathology. Genetic ablation of Il24 or the receptor Il20rb in keratinocytes alleviated allergic inflammation and atopic march. Mechanistically, IL-24 acted through its heterodimeric receptors on keratinocytes and augmented the production of IL-33, which in turn aggravated type 2 immunity and AD-like skin conditions. Overall, these findings establish IL-24 as a critical factor for onset and progression of AD and a compelling therapeutic target.
Keywords: IL-24; MRSA; allergic inflammation; atopic dermatitis; keratinocytes.
© The Author(s) 2024. Published by Oxford University Press on behalf of Higher Education Press.
Conflict of interest statement
Authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

