γδ T cell profiling in a cohort of preterm infants reveals elevated frequencies of CD83+ γδ T cells in sepsis
- PMID: 38753245
- PMCID: PMC11098939
- DOI: 10.1084/jem.20231987
γδ T cell profiling in a cohort of preterm infants reveals elevated frequencies of CD83+ γδ T cells in sepsis
Abstract
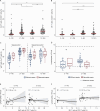
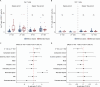
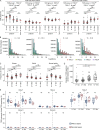
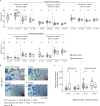
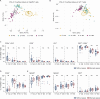
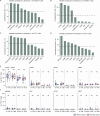
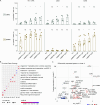
Preterm infants are at high risk of developing neonatal sepsis. γδ T cells are thought to be an important set of effector cells in neonates. Here, γδ T cells were investigated in a longitudinal cohort of preterm neonates using next-generation sequencing, flow cytometry, and functional assays. During the first year of life, the Vγ9Vδ2 T cell subset showed dynamic phenotypic changes and elevated levels of fetal-derived Vγ9Vδ2 T cells were evident in infants with sepsis. Single-cell transcriptomics identified HLA-DRhiCD83+ γδ T cells in neonatal sepsis, which expressed genes related to antigen presentation. In vitro assays showed that CD83 was expressed on activated Vγ9Vδ2 T cells in preterm and term neonates, but not in adults. In contrast, activation of adult Vγ9Vδ2 T cells enhanced CD86 expression, which was presumably the key receptor to induce CD4 T cell proliferation. Together, we provide a map of the maturation of γδ T cells after preterm birth and highlight their phenotypic diversity in infections.
© 2024 Leon-Lara et al.
Conflict of interest statement
Disclosures: S. Pirr reported grants from Deutsche Forschungsgemeinschaft and grants from the German Ministry of Education and Research outside the submitted work. No other disclosures were reported.
Figures




References
-
- Barisa, M., Kramer A.M., Majani Y., Moulding D., Saraiva L., Bajaj-Elliott M., Anderson J., and Gustafsson K.. 2017. E. coli promotes human Vγ9Vδ2 T cell transition from cytokine-producing bactericidal effectors to professional phagocytic killers in a TCR-dependent manner. Sci. Rep. 7:2805. 10.1038/s41598-017-02886-8 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

