Conserved and divergent DNA recognition specificities and functions of R2 retrotransposon N-terminal domains
- PMID: 38753487
- PMCID: PMC11204384
- DOI: 10.1016/j.celrep.2024.114239
Conserved and divergent DNA recognition specificities and functions of R2 retrotransposon N-terminal domains
Abstract
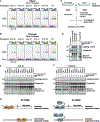
R2 non-long terminal repeat (non-LTR) retrotransposons are among the most extensively distributed mobile genetic elements in multicellular eukaryotes and show promise for applications in transgene supplementation of the human genome. They insert new gene copies into a conserved site in 28S ribosomal DNA with exquisite specificity. R2 clades are defined by the number of zinc fingers (ZFs) at the N terminus of the retrotransposon-encoded protein, postulated to additively confer DNA site specificity. Here, we illuminate general principles of DNA recognition by R2 N-terminal domains across and between clades, with extensive, specific recognition requiring only one or two compact domains. DNA-binding and protection assays demonstrate broadly shared as well as clade-specific DNA interactions. Gene insertion assays in cells identify the N-terminal domains sufficient for target-site insertion and reveal roles in second-strand cleavage or synthesis for clade-specific ZFs. Our results have implications for understanding evolutionary diversification of non-LTR retrotransposon insertion mechanisms and the design of retrotransposon-based gene therapies.
Keywords: CP: Molecular biology; DNA-binding specificity; Myb domain; R2 retrotransposon; gene insertion; gene therapy; genome engineering; non-LTR retrotransposon; protein-DNA interaction; zinc finger.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.V.T. and K.C. are listed inventors on patent applications filed by University of California, Berkeley, related to the transgene insertion technology platform. B.V.T. and K.C. have equity options in Addition Therapeutics, which licensed the University of California, Berkeley technology. K.C. is a consultant and board member of Addition Therapeutics but does not receive personal compensation for these roles.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

