How total mRNA influences cell growth
- PMID: 38753514
- PMCID: PMC11126920
- DOI: 10.1073/pnas.2400679121
How total mRNA influences cell growth
Abstract
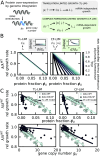
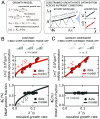
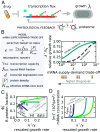
Experimental observations tracing back to the 1960s imply that ribosome quantities play a prominent role in determining a cell's growth. Nevertheless, in biologically relevant scenarios, growth can also be influenced by the levels of mRNA and RNA polymerase. Here, we construct a quantitative model of biosynthesis providing testable scenarios for these situations. The model explores a theoretically motivated regime where RNA polymerases compete for genes and ribosomes for transcripts and gives general expressions relating growth rate, mRNA concentrations, ribosome, and RNA polymerase levels. On general grounds, the model predicts how the fraction of ribosomes in the proteome depends on total mRNA concentration and inspects an underexplored regime in which the trade-off between transcript levels and ribosome abundances sets the cellular growth rate. In particular, we show that the model predicts and clarifies three important experimental observations, in budding yeast and Escherichia coli bacteria: i) that the growth-rate cost of unneeded protein expression can be affected by mRNA levels, ii) that resource optimization leads to decreasing trends in mRNA levels at slow growth, and iii) that ribosome allocation may increase, stay constant, or decrease, in response to transcription-inhibiting antibiotics. Since the data indicate that a regime of joint limitation may apply in physiological conditions and not only to perturbations, we speculate that this regime is likely self-imposed.
Keywords: biosynthesis; cell growth; mRNA; transcription; translation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

Similar articles
-
A counting-strategy together with a spatial structured model describes RNA polymerase and ribosome availability in Escherichia coli.Metab Eng. 2021 Sep;67:145-152. doi: 10.1016/j.ymben.2021.06.006. Epub 2021 Jun 23. Metab Eng. 2021. PMID: 34174424
-
Synergistic Regulation of Transcription and Translation in Escherichia coli Revealed by Codirectional Increases in mRNA Concentration and Translation Efficiency.Microbiol Spectr. 2022 Feb 23;10(1):e0204121. doi: 10.1128/spectrum.02041-21. Epub 2022 Feb 9. Microbiol Spectr. 2022. PMID: 35138139 Free PMC article.
-
Algorithms for ribosome traffic engineering and their potential in improving host cells' titer and growth rate.Sci Rep. 2020 Dec 3;10(1):21202. doi: 10.1038/s41598-020-78260-y. Sci Rep. 2020. PMID: 33273552 Free PMC article.
-
Polymerase stalling during replication, transcription and translation.Curr Biol. 2014 May 19;24(10):R445-52. doi: 10.1016/j.cub.2014.03.060. Curr Biol. 2014. PMID: 24845677 Review.
-
RNA Polymerase's Relationship with the Ribosome: Not So Physical, Most of the Time.J Mol Biol. 2020 Jun 26;432(14):3981-3986. doi: 10.1016/j.jmb.2020.03.018. Epub 2020 Mar 19. J Mol Biol. 2020. PMID: 32198117 Free PMC article. Review.
Cited by
-
Single-cell heterogeneity in ribosome content and the consequences for the growth laws.bioRxiv [Preprint]. 2024 Oct 8:2024.04.19.590370. doi: 10.1101/2024.04.19.590370. bioRxiv. 2024. PMID: 38895328 Free PMC article. Preprint.
-
The Product neutrality function defining genetic interactions emerges from mechanistic models of cell growth.Elife. 2025 Sep 2;14:RP105265. doi: 10.7554/eLife.105265. Elife. 2025. PMID: 40891677 Free PMC article.
References
-
- Neidhardt F. C., Magasanik B., Studies on the role of ribonucleic acid in the growth of bacteria. Biochim. Biophys. Acta 42, 99–116 (1960). - PubMed
-
- Koch A. L., Why can’t a cell grow infinitely fast? Can. J. Microbiol. 34, 421–426 (1988). - PubMed
-
- Bremer H., Dennis P. P., Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus 3 (2008). - PubMed

