Leishmania Ribosomal Protein (RP) paralogous genes compensate each other's expression maintaining protein native levels
- PMID: 38753846
- PMCID: PMC11098316
- DOI: 10.1371/journal.pone.0292152
Leishmania Ribosomal Protein (RP) paralogous genes compensate each other's expression maintaining protein native levels
Abstract
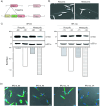
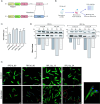
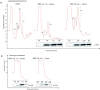
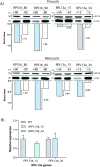
In the protozoan parasite Leishmania, most genes encoding for ribosomal proteins (RPs) are present as two or more copies in the genome. However, their untranslated regions (UTRs) are predominantly divergent and might be associated with a distinct regulation of the expression of paralogous genes. Herein, we investigated the expression profiles of two RPs (S16 and L13a) encoded by duplicated genes in Leishmania major. The genes encoding for the S16 protein possess identical coding sequences (CDSs) and divergent UTRs, whereas the CDSs of L13a diverge by two amino acids and by their UTRs. Using CRISPR/Cas9 genome editing, we generated knockout (Δ) and endogenously tagged transfectants for each paralog of L13a and S16 genes. Combining tagged and Δ cell lines we found evidence of differential expression of both RPS16 and RPL13a isoforms throughout parasite development, with one isoform consistently more abundant than its respective copy. In addition, compensatory expression was observed for each paralog upon deletion of the corresponding isoform, suggesting functional conservation between these proteins. This differential expression pattern relates to post-translational processes, given compensation occurs at the level of the protein, with no alterations detected at transcript level. Ribosomal profiles for RPL13a indicate a standard behavior for these paralogues suggestive of interaction with heavy RNA-protein complexes, as already reported for other RPs in trypanosomatids. We identified paralog-specific bound to their 3'UTRs which may be influential in regulating paralog expression. In support, we identified conserved cis-elements within the 3'UTRs of RPS16 and RPL13a; cis-elements exclusive to the UTR of the more abundant paralog or to the less abundant ones were identified.
Copyright: © 2024 Borges et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Generation of 3'UTR knockout cell lines by CRISPR/Cas9-mediated genome editing.Methods Enzymol. 2021;655:427-457. doi: 10.1016/bs.mie.2021.03.014. Epub 2021 May 28. Methods Enzymol. 2021. PMID: 34183132
-
Translational machinery of the chaetognath Spadella cephaloptera: a transcriptomic approach to the analysis of cytosolic ribosomal protein genes and their expression.BMC Evol Biol. 2007 Aug 28;7:146. doi: 10.1186/1471-2148-7-146. BMC Evol Biol. 2007. PMID: 17725830 Free PMC article.
-
Application of CRISPR/Cas9-Based Reverse Genetics in Leishmania braziliensis: Conserved Roles for HSP100 and HSP23.Genes (Basel). 2020 Sep 30;11(10):1159. doi: 10.3390/genes11101159. Genes (Basel). 2020. PMID: 33007987 Free PMC article.
-
Bacterial 3'UTRs: A Useful Resource in Post-transcriptional Regulation.Front Mol Biosci. 2021 Jan 8;7:617633. doi: 10.3389/fmolb.2020.617633. eCollection 2020. Front Mol Biosci. 2021. PMID: 33490108 Free PMC article. Review.
-
Characterization of the multigene family encoding the mouse S16 ribosomal protein: strategy for distinguishing an expressed gene from its processed pseudogene counterparts by an analysis of total genomic DNA.Mol Cell Biol. 1985 Dec;5(12):3560-76. doi: 10.1128/mcb.5.12.3560-3576.1985. Mol Cell Biol. 1985. PMID: 3915781 Free PMC article. Review.
Cited by
-
Ribosome Structural Changes Dynamically Affect Ribosome Function.Int J Mol Sci. 2024 Oct 17;25(20):11186. doi: 10.3390/ijms252011186. Int J Mol Sci. 2024. PMID: 39456968 Free PMC article. Review.
-
Beyond the ORF: Paralog-specific regulation of RPS7/eS7 mRNAs via 3'-UTRs and promoter sequences.PLoS One. 2025 May 30;20(5):e0324525. doi: 10.1371/journal.pone.0324525. eCollection 2025. PLoS One. 2025. PMID: 40445952 Free PMC article.
References
-
- Soto M, Requena JM, Garcia M, Gomez LC, Navarrete I, Alonso C. Genomic organization and expression of two independent gene arrays coding for two antigenic acidic ribosomal proteins of Leishmania. J Biol Chem. 1993;268(29):21835–43. - PubMed
-
- Sim EU-H, Er C-M. Extra-Ribosomal Functions of the Ribosomal Protein, RPS3 as Predicted by In Silico Analysis. Borneo J Resour Sci Technol. 1970;4(2):62–9.
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

