Nuclear export is a limiting factor in eukaryotic mRNA metabolism
- PMID: 38753883
- PMCID: PMC11135743
- DOI: 10.1371/journal.pcbi.1012059
Nuclear export is a limiting factor in eukaryotic mRNA metabolism
Abstract
The eukaryotic mRNA life cycle includes transcription, nuclear mRNA export and degradation. To quantify all these processes simultaneously, we perform thiol-linked alkylation after metabolic labeling of RNA with 4-thiouridine (4sU), followed by sequencing of RNA (SLAM-seq) in the nuclear and cytosolic compartments of human cancer cells. We develop a model that reliably quantifies mRNA-specific synthesis, nuclear export, and nuclear and cytosolic degradation rates on a genome-wide scale. We find that nuclear degradation of polyadenylated mRNA is negligible and nuclear mRNA export is slow, while cytosolic mRNA degradation is comparatively fast. Consequently, an mRNA molecule generally spends most of its life in the nucleus. We also observe large differences in the nuclear export rates of different 3'UTR transcript isoforms. Furthermore, we identify genes whose expression is abruptly induced upon metabolic labeling. These transcripts are exported substantially faster than average mRNAs, suggesting the existence of alternative export pathways. Our results highlight nuclear mRNA export as a limiting factor in mRNA metabolism and gene regulation.
Copyright: © 2024 Müller et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
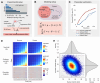
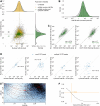


References
-
- Boehringer A, Bowser R. RNA Nucleocytoplasmic Transport Defects in Neurodegenerative Diseases. In: Sattler R, Donnelly CJ, editors. RNA Metabolism in Neurodegenerative Diseases. Advances in Neurobiology. Cham: Springer International Publishing; 2018. p. 85–101. Available from: 10.1007/978-3-319-89689-2_4 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

