TDP-43 proteinopathy in ALS is triggered by loss of ASRGL1 and associated with HML-2 expression
- PMID: 38755145
- PMCID: PMC11099023
- DOI: 10.1038/s41467-024-48488-7
TDP-43 proteinopathy in ALS is triggered by loss of ASRGL1 and associated with HML-2 expression
Abstract
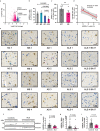
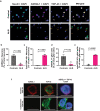
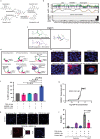
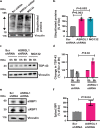
TAR DNA-binding protein 43 (TDP-43) proteinopathy in brain cells is the hallmark of amyotrophic lateral sclerosis (ALS) but its cause remains elusive. Asparaginase-like-1 protein (ASRGL1) cleaves isoaspartates, which alter protein folding and susceptibility to proteolysis. ASRGL1 gene harbors a copy of the human endogenous retrovirus HML-2, whose overexpression contributes to ALS pathogenesis. Here we show that ASRGL1 expression was diminished in ALS brain samples by RNA sequencing, immunohistochemistry, and western blotting. TDP-43 and ASRGL1 colocalized in neurons but, in the absence of ASRGL1, TDP-43 aggregated in the cytoplasm. TDP-43 was found to be prone to isoaspartate formation and a substrate for ASRGL1. ASRGL1 silencing triggered accumulation of misfolded, fragmented, phosphorylated and mislocalized TDP-43 in cultured neurons and motor cortex of female mice. Overexpression of ASRGL1 restored neuronal viability. Overexpression of HML-2 led to ASRGL1 silencing. Loss of ASRGL1 leading to TDP-43 aggregation may be a critical mechanism in ALS pathophysiology.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

