ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-2024 update
- PMID: 38755420
- PMCID: PMC11217004
- DOI: 10.1038/s41375-024-02267-x
ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-2024 update
Abstract
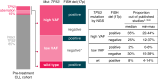
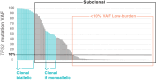
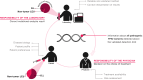
In chronic lymphocytic leukemia (CLL), analysis of TP53 aberrations (deletion and/or mutation) is a crucial part of treatment decision-making algorithms. Technological and treatment advances have resulted in the need for an update of the last recommendations for TP53 analysis in CLL, published by ERIC, the European Research Initiative on CLL, in 2018. Based on the current knowledge of the relevance of low-burden TP53-mutated clones, a specific variant allele frequency (VAF) cut-off for reporting TP53 mutations is no longer recommended, but instead, the need for thorough method validation by the reporting laboratory is emphasized. The result of TP53 analyses should always be interpreted within the context of available laboratory and clinical information, treatment indication, and therapeutic options. Methodological aspects of introducing next-generation sequencing (NGS) in routine practice are discussed with a focus on reliable detection of low-burden clones. Furthermore, potential interpretation challenges are presented, and a simplified algorithm for the classification of TP53 variants in CLL is provided, representing a consensus based on previously published guidelines. Finally, the reporting requirements are highlighted, including a template for clinical reports of TP53 aberrations. These recommendations are intended to assist diagnosticians in the correct assessment of TP53 mutation status, but also physicians in the appropriate understanding of the lab reports, thus decreasing the risk of misinterpretation and incorrect management of patients in routine practice whilst also leading to improved stratification of patients with CLL in clinical trials.
© 2024. The Author(s).
Conflict of interest statement
SPav has received honoraria from AstraZeneca. PB has received honoraria from Abbvie, Gilead and Janssen, and research funding from Gilead. ET has received honoraria from Abbvie, AstraZeneca, BeiGene, Janssen and Hoffmann-La Roche, and research support from Abbvie, Roche and Gilead. DR has received honoraria from AbbVie, AstraZeneca, BeiGene, BMS, Janssen and Lilly, research grants from AbbVie, AstraZeneca and Janssen, and travel grants from AstraZeneca and Janssen. AK has received research funding from BMS, Astra Zeneca, Janssen, Abbvie and Roche Genentech, and compensation as a member of the scientific advisory board from BMS, Astra Zeneca, Janssen, Abbvie, Roche Genentec and LAVA. CUN has received research funding and/or consultancy fees from Abbvie, AstraZeneca, Beigene, Janssen, Genmab, Lilly, MSD, CSL Behring, Takeda and Octapharma. FD has received honoraria from Janssen and AstraZeneca. GG has received compensation as a member of the scientific advisory board from Abbvie, Astra Zeneca, BeiGene, Incyte, Janssen and Lilly, and Speaker’s Bureau honoraria from Abbvie, BeiGene, Astra Zeneca and Janssen. SS has received compensation as a member of the scientific advisory board, research support, travel support and speaker fees from AbbVie, Acerta, Amgen, AstraZeneca, BeiGene, BMS, Celgene, Gilead, GSK, Hoffmann-La Roche, Infinity, Janssen, Lilly, Novartis, Sunesis, and Verastem. RR has received honoraria from AbbVie, AstraZeneca, Janssen, Illumina, and Roche. KS has received research funding, honoraria and/or consultancy fees from Abbvie, AstraZeneca, Janssen, Lilly and Roche. PG has received honoraria from AbbVie, Astrazeneca, BeiGene, BMS, Galapagos, Janssen, Lilly/Loxo Oncology, MSD and Roche, and research funding from AbbVie, Astrazeneca, BMS and Janssen.
Figures
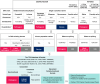
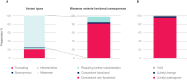
References
-
- Zenz T, Habe S, Denzel T, Mohr J, Winkler D, Buhler A, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory CLL: dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114:2589–97. - PubMed
-
- Rossi D, Spina V, Deambrogi C, Rasi S, Laurenti L, Stamatopoulos K, et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood. 2011;117:3391–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

