Population dynamics of potentially harmful haplotypes: a pedigree analysis
- PMID: 38755557
- PMCID: PMC11097446
- DOI: 10.1186/s12864-024-10407-x
Population dynamics of potentially harmful haplotypes: a pedigree analysis
Abstract
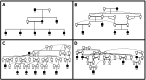
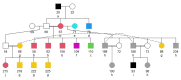
Background: The identification of low-frequency haplotypes, never observed in homozygous state in a population, is considered informative on the presence of potentially harmful alleles (candidate alleles), putatively involved in inbreeding depression. Although identification of candidate alleles is challenging, studies analyzing the dynamics of potentially harmful alleles are lacking. A pedigree of the highly endangered Gochu Asturcelta pig breed, including 471 individuals belonging to 51 different families with at least 5 offspring each, was genotyped using the Axiom PigHDv1 Array (658,692 SNPs). Analyses were carried out on four different cohorts defined according to pedigree depth and at the whole population (WP) level.
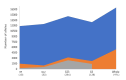
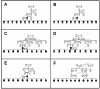
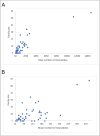
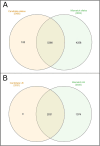
Results: The 4,470 Linkage Blocks (LB) identified in the Base Population (10 individuals), gathered a total of 16,981 alleles in the WP. Up to 5,466 (32%) haplotypes were statistically considered candidate alleles, 3,995 of them (73%) having one copy only. The number of alleles and candidate alleles varied across cohorts according to sample size. Up to 4,610 of the alleles identified in the WP (27% of the total) were present in one cohort only. Parentage analysis identified a total of 67,742 parent-offspring incompatibilities. The number of mismatches varied according to family size. Parent-offspring inconsistencies were identified in 98.2% of the candidate alleles and 100% of the LB in which they were located. Segregation analyses informed that most potential candidate alleles appeared de novo in the pedigree. Only 17 candidate alleles were identified in the boar, sow, and paternal and maternal grandparents and were considered segregants.
Conclusions: Our results suggest that neither mutation nor recombination are the major forces causing the apparition of candidate alleles. Their occurrence is more likely caused by Allele-Drop-In events due to SNP calling errors. New alleles appear when wrongly called SNPs are used to construct haplotypes. The presence of candidate alleles in either parents or grandparents of the carrier individuals does not ensure that they are true alleles. Minimum Allele Frequency thresholds may remove informative alleles. Only fully segregant candidate alleles should be considered potentially harmful alleles. A set of 16 candidate genes, potentially involved in inbreeding depression, is described.
Keywords: Allele-Drop-In events; Linkage blocks; Pedigree variation; Population genomics; Potentially harmful alleles.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. - PubMed
-
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of Haplotype blocks in the Human Genome. Science. 2002;296(5576):2225–9. - PubMed
-
- Utsunomiya YT, Milanesi M, Utsunomiya ATH, Ajmone-Marsan P, Garcia JF. GHap: an R package for genome-wide haplotyping. Bioinformatics. 2016;32(18):2861–2. - PubMed
MeSH terms
Grants and funding
- PID2019-103951RB/AEI/10.13039/501100011033/Agencia Estatal de Investigación
- PID2019-103951RB/AEI/10.13039/501100011033/Agencia Estatal de Investigación
- PID2019-103951RB/AEI/10.13039/501100011033/Agencia Estatal de Investigación
- PID2019-103951RB/AEI/10.13039/501100011033/Agencia Estatal de Investigación
LinkOut - more resources
Full Text Sources

