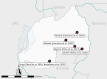
Expansion of artemisinin partial resistance mutations and lack of histidine rich protein-2 and -3 deletions in Plasmodium falciparum infections from Rukara, Rwanda
- PMID: 38755607
- PMCID: PMC11100144
- DOI: 10.1186/s12936-024-04981-4
Expansion of artemisinin partial resistance mutations and lack of histidine rich protein-2 and -3 deletions in Plasmodium falciparum infections from Rukara, Rwanda
Abstract
Background: Emerging artemisinin partial resistance and diagnostic resistance are a threat to malaria control in Africa. Plasmodium falciparum kelch13 (k13) propeller-domain mutations that confer artemisinin partial resistance have emerged in Africa. k13-561H was initially described at a frequency of 7.4% from Masaka in 2014-2015, but not present in nearby Rukara. By 2018, 19.6% of isolates in Masaka and 22% of isolates in Rukara contained the mutation. Longitudinal monitoring is essential to inform control efforts. In Rukara, an assessment was conducted to evaluate recent k13-561H prevalence changes, as well as other key mutations. Prevalence of hrp2/3 deletions was also assessed.
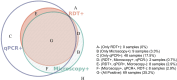
Methods: Samples collected in Rukara in 2021 were genotyped for key artemisinin and partner drug resistance mutations using molecular inversion probe assays and for hrp2/3 deletions using qPCR.
Results: Clinically validated k13 artemisinin partial resistance mutations continue to increase in prevalence with the overall level of mutant infections reaching 32% in Rwanda. The increase appears to be due to the rapid emergence of k13-675V (6.4%, 6/94 infections), previously not observed, rather than continued expansion of 561H (23.5% 20/85). Mutations to partner drugs and other anti-malarials were variable, with high levels of multidrug resistance 1 (mdr1) N86 (95.5%) associated with lumefantrine decreased susceptibility and dihydrofolate reductase (dhfr) 164L (24.7%) associated with a high level of antifolate resistance, but low levels of amodiaquine resistance polymorphisms with chloroquine resistance transporter (crt) 76T: at 6.1% prevalence. No hrp2 or hrp3 gene deletions associated with diagnostic resistance were found.
Conclusions: Increasing prevalence of artemisinin partial resistance due to k13-561H and the rapid expansion of k13-675V is concerning for the longevity of artemisinin effectiveness in the region. False negative RDT results do not appear to be an issue with no hrp2 or hpr3 deletions detected. Continued molecular surveillance in this region and surrounding areas is needed to follow artemisinin partial resistance and provide early detection of partner drug resistance, which would likely compromise control and increase malaria morbidity and mortality in East Africa.
Keywords: Plasmodium falciparum; Artemisinin; Drug resistance; K13; Malaria; R561H; Rukara; Rwanda; kelch13.
© 2024. The Author(s).
Conflict of interest statement
The authors have no competing interests to report. The funder had no role in the implementation and interpretation of the project.
Figures


Update of
-
Expansion of Artemisinin Partial Resistance Mutations and Lack of Histidine Rich Protein-2 and -3 Deletions in Plasmodium falciparum infections from Rukara, Rwanda.medRxiv [Preprint]. 2023 Dec 18:2023.12.17.23300081. doi: 10.1101/2023.12.17.23300081. medRxiv. 2023. Update in: Malar J. 2024 May 16;23(1):150. doi: 10.1186/s12936-024-04981-4. PMID: 38196592 Free PMC article. Updated. Preprint.
References
-
- WHO . World malaria report 2023. Geneva: World Health Organization; 2023.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

