RICTOR/mTORC2 downregulation in BRAFV600E melanoma cells promotes resistance to BRAF/MEK inhibition
- PMID: 38755661
- PMCID: PMC11097536
- DOI: 10.1186/s12943-024-02010-1
RICTOR/mTORC2 downregulation in BRAFV600E melanoma cells promotes resistance to BRAF/MEK inhibition
Abstract
Background: The main drawback of BRAF/MEK inhibitors (BRAF/MEKi)-based targeted therapy in the management of BRAF-mutated cutaneous metastatic melanoma (MM) is the development of therapeutic resistance. We aimed to assess in this context the role of mTORC2, a signaling complex defined by the presence of the essential RICTOR subunit, regarded as an oncogenic driver in several tumor types, including MM.
Methods: After analyzing The Cancer Genome Atlas MM patients' database to explore both overall survival and molecular signatures as a function of intra-tumor RICTOR levels, we investigated the effects of RICTOR downregulation in BRAFV600E MM cell lines on their response to BRAF/MEKi. We performed proteomic screening to identify proteins modulated by changes in RICTOR expression, and Seahorse analysis to evaluate the effects of RICTOR depletion on mitochondrial respiration. The combination of BRAFi with drugs targeting proteins and processes emerged in the proteomic screening was carried out on RICTOR-deficient cells in vitro and in a xenograft setting in vivo.
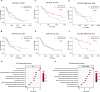
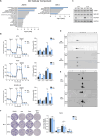
Results: Low RICTOR levels in BRAF-mutated MM correlate with a worse clinical outcome. Gene Set Enrichment Analysis of low-RICTOR tumors display gene signatures suggestive of activation of the mitochondrial Electron Transport Chain (ETC) energy production. RICTOR-deficient BRAFV600E cells are intrinsically tolerant to BRAF/MEKi and anticipate the onset of resistance to BRAFi upon prolonged drug exposure. Moreover, in drug-naïve cells we observed a decline in RICTOR expression shortly after BRAFi exposure. In RICTOR-depleted cells, both mitochondrial respiration and expression of nicotinamide phosphoribosyltransferase (NAMPT) are enhanced, and their pharmacological inhibition restores sensitivity to BRAFi.
Conclusions: Our work unveils an unforeseen tumor-suppressing role for mTORC2 in the early adaptation phase of BRAFV600E melanoma cells to targeted therapy and identifies the NAMPT-ETC axis as a potential therapeutic vulnerability of low RICTOR tumors. Importantly, our findings indicate that the evaluation of intra-tumor RICTOR levels has a prognostic value in metastatic melanoma and may help to guide therapeutic strategies in a personalized manner.
Keywords: BRAFV600E melanoma; Drug resistance; Mitochondrial metabolism; NAMPT; RICTOR; Targeted therapy; mTORC2.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Tangella LP, Clark ME, Gray ES. Resistance mechanisms to targeted therapy in BRAF-mutant melanoma - A mini review. Biochimica et Biophysica Acta - General Subjects. Volume 1865. Elsevier B.V.; 2021. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

