Screening of key immune - related gene in Parkinson ' s disease based on WGCNA and machine learning
- PMID: 38755717
- PMCID: PMC11103055
- DOI: 10.11817/j.issn.1672-7347.2024.230307
Screening of key immune - related gene in Parkinson ' s disease based on WGCNA and machine learning
Abstract
Objectives: Abnormal immune system activation and inflammation are crucial in causing Parkinson's disease. However, we still don't fully understand how certain immune-related genes contribute to the disease's development and progression. This study aims to screen key immune-related gene in Parkinson's disease based on weighted gene co-expression network analysis (WGCNA) and machine learning.
Methods: This study downloaded the gene chip data from the Gene Expression Omnibus (GEO) database, and used WGCNA to screen out important gene modules related to Parkinson's disease. Genes from important modules were exported and a Venn diagram of important Parkinson's disease-related genes and immune-related genes was drawn to screen out immune related genes of Parkinson's disease. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to analyze the the functions of immune-related genes and signaling pathways involved. Immune cell infiltration analysis was performed using the CIBERSORT package of R language. Using bioinformatics method and 3 machine learning methods [least absolute shrinkage and selection operator (LASSO) regression, random forest (RF), and support vector machine (SVM)], the immune-related genes of Parkinson's disease were further screened. A Venn diagram of differentially expressed genes screened using the 4 methods was drawn with the intersection gene being hub nodes (hub) gene. The downstream proteins of the Parkinson's disease hub gene was identified through the STRING database and a protein-protein interaction network diagram was drawn.
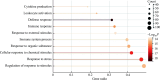
Results: A total of 218 immune genes related to Parkinson's disease were identified, including 45 upregulated genes and 50 downregulated genes. Enrichment analysis showed that the 218 genes were mainly enriched in immune system response to foreign substances and viral infection pathways. The results of immune infiltration analysis showed that the infiltration percentages of CD4+ T cells, NK cells, CD8+ T cells, and B cells were higher in the samples of Parkinson's disease patients, while resting NK cells and resting CD4+ T cells were significantly infiltrated in the samples of Parkinson's disease patients. ANK1 was screened out as the hub gene. The analysis of the protein-protein interaction network showed that the ANK1 translated and expressed 11 proteins which mainly participated in functions such as signal transduction, iron homeostasis regulation, and immune system activation.
Conclusions: This study identifies the Parkinson's disease immune-related key gene ANK1 via WGCNA and machine learning methods, suggesting its potential as a candidate therapeutic target for Parkinson's disease.
目的: 在帕金森病的发病过程中,免疫系统的异常激活和炎症反应起着重要作用。然而,目前对于免疫相关关键基因在帕金森病发生和发展中的具体作用和作用机制的了解仍然有限。本研究旨在通过加权基因共表达网络分析(weighted gene co-expression network analysis,WGCNA)和机器学习筛选帕金森病免疫相关关键基因。方法: 从基因表达综合(Gene Expression Omnibus,GEO)数据库下载基因芯片数据,采用WGCNA筛选出与帕金森病相关的重要基因模块;将重要模块中的基因导出,绘制帕金森病重要相关基因与免疫相关基因的韦恩图,从而筛选出帕金森病免疫相关基因。采用基因本体(gene ontology,GO)分析和京都基因和基因组百科全书(Kyoto Encyclopedia of Genes and Genomes,KEGG)深入分析免疫相关基因的功能及参与的信号通路。通过R语言的CIBERSORT包进行免疫细胞浸润分析。采用生物信息学方法和3种机器学习方法[最小绝对收缩和选择算子(least absolute shrinkage and selection operator,LASSO)回归、随机森林(random forest,RF)和支持向量机(support vector machine,SVM)]对筛选出的帕金森病免疫相关基因进行进一步筛选研究,绘制4种方法筛选的差异表达基因的韦恩图,筛选交集基因即中心节点(hub node,hub)基因。通过STRING数据库搜索帕金森病hub基因的下游蛋白质,绘制蛋白质互作网络图。结果: 筛选出帕金森病重要模块基因中与免疫相关的基因218个,其中45个为上调基因,50个为下调基因。富集分析结果显示218个基因主要在免疫系统对外来物反应和病毒感染通路富集。免疫浸润分析结果表明,CD4+ T细胞、NK细胞、CD8+ T细胞、B细胞在帕金森病患者样本中的浸润百分率较高,静息NK细胞、静息CD4+ T细胞在帕金森病患者样本中显著浸润。4种方法筛选出的hub基因为ANK1基因。交集基因蛋白质互作网络分析结果显示,ANK1基因翻译表达的11个蛋白质主要参与信号转导、铁稳态调节及免疫系统激活等功能。结论: 通过WGCNA和机器学习方法,筛选出帕金森病免疫相关关键基因ANK1,该基因可能成为帕金森病诊断和治疗的候选靶点。.
Keywords: ANK1 gene; Parkinson’s disease; immunity; machine learning; weighted gene co-expression network analysis.
Conflict of interest statement
作者声称无任何利益冲突。
Figures













Similar articles
-
[Exploration of key ferroptosis-related genes as therapeutic targets for sepsis based on bioinformatics and the depiction of their immune profiles characterization].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2024 Oct;36(10):1025-1032. doi: 10.3760/cma.j.cn121430-20240524-00457. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2024. PMID: 39586719 Chinese.
-
Identifying the hub gene and immune infiltration of Parkinson's disease using bioinformatical methods.Brain Res. 2022 Jun 15;1785:147879. doi: 10.1016/j.brainres.2022.147879. Epub 2022 Mar 10. Brain Res. 2022. PMID: 35278479
-
Exploration of the shared diagnostic genes and mechanisms between periodontitis and primary Sjögren's syndrome by integrated comprehensive bioinformatics analysis and machine learning.Int Immunopharmacol. 2024 Nov 15;141:112899. doi: 10.1016/j.intimp.2024.112899. Epub 2024 Aug 13. Int Immunopharmacol. 2024. PMID: 39142001
-
Bioinformatics and machine learning approaches reveal key genes and underlying molecular mechanisms of atherosclerosis: A review.Medicine (Baltimore). 2024 Aug 2;103(31):e38744. doi: 10.1097/MD.0000000000038744. Medicine (Baltimore). 2024. PMID: 39093811 Free PMC article. Review.
-
The role of AI and machine learning in the diagnosis of Parkinson's disease and atypical parkinsonisms.Parkinsonism Relat Disord. 2024 Sep;126:106986. doi: 10.1016/j.parkreldis.2024.106986. Epub 2024 May 3. Parkinsonism Relat Disord. 2024. PMID: 38724317 Review.
References
-
- WHO . Launch of WHO’s Parkinson disease technical brief [Z]. World Health Organization, 2022.
-
- 汤琳琳, 尤浩军, 雷静. 帕金森病的病理性痛机制与治疗进展[J]. 生理学报, 2023, 75(4): 595-603. 10.13294/japs.2023.0042. - DOI - PubMed
- TANG Linlin, YOU Haojun, LEI Jing. Pathological pain mechanism and treatment progress of Parkinson’s disease[J]. Acta Physiologica Sinica, 2023, 75(4): 595-603. 10.13294/japs.2023.0042. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

