An Optimized and Advanced Algorithm for the Quantification of Immunohistochemical Biomarkers in Keratinocytes
- PMID: 38756235
- PMCID: PMC11097113
- DOI: 10.1016/j.xjidi.2024.100270
An Optimized and Advanced Algorithm for the Quantification of Immunohistochemical Biomarkers in Keratinocytes
Abstract
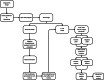
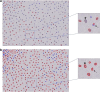
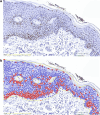
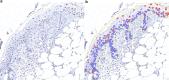
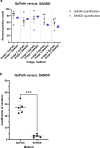
Advancements in pathology have given rise to software applications intended to minimize human error and improve efficacy of image analysis. Still, the subjectivity of image quantification performed manually and the limitations of the most ubiquitous tissue stain analysis software requiring parameters tuned by the observer, reveal the need for a highly accurate, automated nuclear quantification software specific to immunohistochemistry, with improved precision and efficiency compared with the methods currently in use. We present a method for the quantification of immunohistochemical biomarkers in keratinocyte nuclei proposed to overcome these limitations, contributing sensitive shape-focused segmentation, accurate nuclear detection, and automated device-independent color assessment, without observer-dependent analysis parameters.
Keywords: Automated digital pathology; Biomarker; Epidermal structures; Immunohistochemistry; Quantification.
© 2024 Published by Elsevier Inc. on behalf of the Society for Investigative Dermatology.
Figures






References
-
- Akakin H, Gokozan H, Otero J, Gurcan M. An adaptive algorithm for detection of multiple-type, positively stained nuclei in IHC images with minimal prior information: application to OLIG2 staining gliomas. Paper presented at: SPIE Medical Imaging. 2015; Olando, FL.
-
- Bradski G. The OpenCV Library. Dr Dobbs J. 2000;25:120–125.
-
- Chen L., Bao J., Huang Q., Sun H. A robust and automated cell counting method in quantification of digital breast cancer immunohistochemistry images. Pol J Pathol. 2019;70:162–173. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

