The sugar donor specificity of plant family 1 glycosyltransferases
- PMID: 38756413
- PMCID: PMC11096472
- DOI: 10.3389/fbioe.2024.1396268
The sugar donor specificity of plant family 1 glycosyltransferases
Abstract
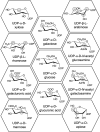
Plant family 1 glycosyltransferases (UGTs) represent a formidable tool to produce valuable natural and novel glycosides. Their regio- and stereo-specific one-step glycosylation mechanism along with their inherent wide acceptor scope are desirable traits in biotechnology. However, their donor scope and specificity are not well understood. Since different sugars have different properties in vivo and in vitro, the ability to easily glycodiversify target acceptors is desired, and this depends on our improved understanding of the donor binding site. In the aim to unlock the full potential of UGTs, studies have attempted to elucidate the structure-function relationship governing their donor specificity. These efforts have revealed a complex phenomenon, and general principles valid for multiple enzymes are elusive. Here, we review the studies of UGT donor specificity, and attempt to group the information into key concepts which can help shape future research. We zoom in on the family-defining PSPG motif, on two loop residues reported to interact with the C6 position of the sugar, and on the role of active site arginines in donor specificity. We continue to discuss attempts to alter and expand the donor specificity by enzyme engineering, and finally discuss future research directions.
Keywords: donor specificity; enzyme engineering; glycodiversification; glycosyltransferase; structure-function.
Copyright © 2024 Gharabli and Welner.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
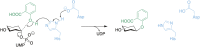



References
-
- Brazier-Hicks M., Offen W. A., Gershater M. C., Revett T. J., Lim E. K., Bowles D. J., et al. (2007). Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. U. S. A. 104, 20238–20243. 10.1073/PNAS.0706421104 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

