High expression of NXPH4 correlates with poor prognosis, metabolic reprogramming, and immune infiltration in colon adenocarcinoma
- PMID: 38756632
- PMCID: PMC11094489
- DOI: 10.21037/jgo-23-956
High expression of NXPH4 correlates with poor prognosis, metabolic reprogramming, and immune infiltration in colon adenocarcinoma
Abstract
Background: Colon adenocarcinoma (COAD) is a prevalent gastrointestinal malignant disease with high mortality rate, and identification of novel prognostic biomarkers and therapeutic targets is urgently needed. Although neurexophilin 4 (NXPH4) has been investigated in several tumors, its role in COAD remains unclear. The aim of this study was to explore the prognostic value and potential functions of NXPH4 in COAD.
Methods: The expression of NXPH4 in COAD were analyzed using The Cancer Genome Atlas (TCGA) and datasets from the Gene Expression Omnibus (GEO) database. The prognostic value of NXPH4 was determined using Kaplan-Meier analysis and Cox regression analysis. To investigate the possible mechanism underlying the role of NXPH4 in COAD, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and gene set enrichment analysis (GSEA) were employed. The correlation between NXPH4 expression and immune cell infiltration levels was examined thorough single-sample gene set enrichment analysis (ssGSEA). Furthermore, the competing endogenous RNA (ceRNA) regulatory network that may be involved in NXPH4 in COAD was predicted and constructed through a variety of databases.
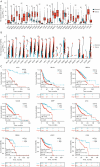
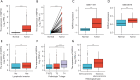
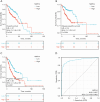
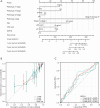
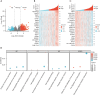
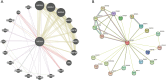
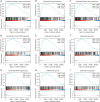
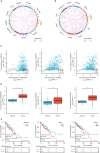
Results: NXPH4 expression was significantly higher in COAD tissue compared with normal colon tissues. Meanwhile, high expression of NXPH4 was associated with poor prognosis in COAD patients. GO-KEGG and GSEA analyses indicated that NXPH4 was associated with glycolysis and hypoxia pathway, and may promote COAD progression and metastasis by modulating metabolic reprogramming. ssGSEA analysis demonstrated that NXPH4 expression also associated with immune infiltration. Furthermore, we identified various microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) as upstream regulators of NXPH4 in COAD.
Conclusions: The present study revealed that high expression of NXPH4 is associated with tumor progression, metabolic reprogramming, and immune infiltration. These findings suggest that NXPH4 could serve as a reliable prognostic biomarker and a promising therapeutic target in COAD.
Keywords: Neurexophilin 4 (NXPH4); colon adenocarcinoma (COAD); immune infiltration; metabolic reprogramming; prognosis.
2024 Journal of Gastrointestinal Oncology. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-23-956/coif). The authors have no conflicts of interest to declare.
Figures

Similar articles
-
Pancancer analysis of NDUFA4L2 with focused role in tumor progression and metastasis of colon adenocarcinoma.Med Oncol. 2024 Oct 14;41(11):285. doi: 10.1007/s12032-024-02531-1. Med Oncol. 2024. PMID: 39402288
-
Identification of a glycolysis- and lactate-related gene signature for predicting prognosis, immune microenvironment, and drug candidates in colon adenocarcinoma.Front Cell Dev Biol. 2022 Aug 23;10:971992. doi: 10.3389/fcell.2022.971992. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36081904 Free PMC article.
-
High MICAL-L2 expression and its role in the prognosis of colon adenocarcinoma.BMC Cancer. 2022 May 2;22(1):487. doi: 10.1186/s12885-022-09614-0. BMC Cancer. 2022. PMID: 35501725 Free PMC article.
-
The Interferon Gamma-Related Long Noncoding RNA Signature Predicts Prognosis and Indicates Immune Microenvironment Infiltration in Colon Adenocarcinoma.Front Oncol. 2022 Jun 7;12:876660. doi: 10.3389/fonc.2022.876660. eCollection 2022. Front Oncol. 2022. PMID: 35747790 Free PMC article.
-
An Intratumor Heterogeneity-Related Signature for Predicting Prognosis, Immune Landscape, and Chemotherapy Response in Colon Adenocarcinoma.Front Med (Lausanne). 2022 Jul 7;9:925661. doi: 10.3389/fmed.2022.925661. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35872794 Free PMC article.
References
LinkOut - more resources
Full Text Sources
