Precise microbiome engineering using natural and synthetic bacteriophages targeting an artificial bacterial consortium
- PMID: 38756723
- PMCID: PMC11096457
- DOI: 10.3389/fmicb.2024.1403903
Precise microbiome engineering using natural and synthetic bacteriophages targeting an artificial bacterial consortium
Abstract
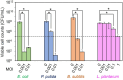
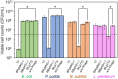
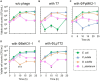
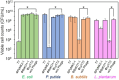
In natural microbiomes, microorganisms interact with each other and exhibit diverse functions. Microbiome engineering, which enables bacterial knockdown, is a promising method to elucidate the functions of targeted bacteria in microbiomes. However, few methods to selectively kill target microorganisms in the microbiome without affecting the growth of nontarget microorganisms are available. In this study, we focused on the host-specific lytic ability of virulent phages and validated their potency for precise microbiome engineering. In an artificial microbiome consisting of Escherichia coli, Pseudomonas putida, Bacillus subtilis, and Lactiplantibacillus plantarum, the addition of bacteriophages infecting their respective host strains specifically reduced the number of these bacteria more than 102 orders. Remarkably, the reduction in target bacteria did not affect the growth of nontarget bacteria, indicating that bacteriophages were effective tools for precise microbiome engineering. Moreover, a virulent derivative of the λ phage was synthesized from prophage DNA in the genome of λ lysogen by in vivo DNA assembly and phage-rebooting techniques, and E. coli-targeted microbiome engineering was achieved. These results propose a novel approach for precise microbiome engineering using bacteriophages, in which virulent phages are synthesized from prophage DNA in lysogenic strains without isolating phages from environmental samples.
Keywords: bacteriophage; microbiome engineering; rebooting; synthetic phage; virulent conversion.
Copyright © 2024 Tanaka, Sugiyama, Sato, Kawaguchi, Honda, Iwaki and Okano.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Programming virulent bacteriophages by developing a multiplex genome engineering method.mBio. 2025 Jun 11;16(6):e0358224. doi: 10.1128/mbio.03582-24. Epub 2025 May 23. mBio. 2025. PMID: 40407328 Free PMC article.
-
Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria.Proc Natl Acad Sci U S A. 2018 Jan 16;115(3):567-572. doi: 10.1073/pnas.1714658115. Epub 2018 Jan 3. Proc Natl Acad Sci U S A. 2018. PMID: 29298913 Free PMC article.
-
Bacteriophages of the Urinary Microbiome.J Bacteriol. 2018 Mar 12;200(7):e00738-17. doi: 10.1128/JB.00738-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29378882 Free PMC article.
-
Engineered bacteriophages: A panacea against pathogenic and drug resistant bacteria.Heliyon. 2024 Jul 9;10(14):e34333. doi: 10.1016/j.heliyon.2024.e34333. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39100447 Free PMC article. Review.
-
Advance on Engineering of Bacteriophages by Synthetic Biology.Infect Drug Resist. 2023 Mar 31;16:1941-1953. doi: 10.2147/IDR.S402962. eCollection 2023. Infect Drug Resist. 2023. PMID: 37025193 Free PMC article. Review.
Cited by
-
Integrating microbial communities into algal biotechnology: a pathway to enhanced commercialization.Front Microbiol. 2025 Apr 1;16:1555579. doi: 10.3389/fmicb.2025.1555579. eCollection 2025. Front Microbiol. 2025. PMID: 40236480 Free PMC article. Review.
-
The Effect of Microbiome-Derived Metabolites in Inflammation-Related Cancer Prevention and Treatment.Biomolecules. 2025 May 8;15(5):688. doi: 10.3390/biom15050688. Biomolecules. 2025. PMID: 40427581 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

