Puerariae lobatae Radix ameliorates chronic kidney disease by reshaping gut microbiota and downregulating Wnt/β‑catenin signaling
- PMID: 38757304
- PMCID: PMC11129539
- DOI: 10.3892/mmr.2024.13241
Puerariae lobatae Radix ameliorates chronic kidney disease by reshaping gut microbiota and downregulating Wnt/β‑catenin signaling
Abstract
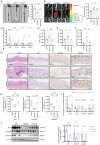
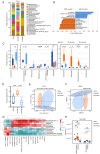
Gut microbiota dysfunction is a key factor affecting chronic kidney disease (CKD) susceptibility. Puerariae lobatae Radix (PLR), a traditional Chinese medicine and food homologous herb, is known to promote the gut microbiota homeostasis; however, its role in renoprotection remains unknown. The present study aimed to investigate the efficacy and potential mechanism of PLR to alleviate CKD. An 8‑week 2% NaCl‑feeding murine model was applied to induce CKD and evaluate the therapeutic effect of PLR supplementary. After gavage for 8 weeks, The medium and high doses of PLR significantly alleviated CKD‑associated creatinine, urine protein increasement and nephritic histopathological injury. Moreover, PLR protected kidney from fibrosis by reducing inflammatory response and downregulating the canonical Wnt/β‑catenin pathway. Furthermore, PLR rescued the gut microbiota dysbiosis and protected against high salt‑induced gut barrier dysfunction. Enrichment of Akkermansia and Bifidobacterium was found after PLR intervention, the relative abundances of which were in positive correlation with normal maintenance of renal histology and function. Next, fecal microbiota transplantation experiment verified that the positive effect of PLR on CKD was, at least partially, exerted through gut microbiota reestablishment and downregulation of the Wnt/β‑catenin pathway. The present study provided evidence for a new function of PLR on kidney protection and put forward a potential therapeutic strategy target for CKD.
Keywords: Puerariae lobatae Radix; Wnt/β‑catenin pathway; chronic kidney disease; fecal microbiota transplantation; gut microbiota; high salt diet.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Gut microbiota-derived melatonin from Puerariae Lobatae Radix-resistant starch supplementation attenuates ischemic stroke injury via a positive microbial co-occurrence pattern.Pharmacol Res. 2023 Apr;190:106714. doi: 10.1016/j.phrs.2023.106714. Epub 2023 Feb 28. Pharmacol Res. 2023. PMID: 36863429
-
Puerariae Lobatae Radix with chuanxiong Rhizoma for treatment of cerebral ischemic stroke by remodeling gut microbiota to regulate the brain-gut barriers.J Nutr Biochem. 2019 Mar;65:101-114. doi: 10.1016/j.jnutbio.2018.12.004. Epub 2018 Dec 21. J Nutr Biochem. 2019. PMID: 30710886
-
Puerariae lobatae Radix Alleviates Pre-Eclampsia by Remodeling Gut Microbiota and Protecting the Gut and Placental Barriers.Nutrients. 2022 Nov 25;14(23):5025. doi: 10.3390/nu14235025. Nutrients. 2022. PMID: 36501055 Free PMC article.
-
[Pathomechanism and treatment of gut microbiota dysbiosis in chronic kidney disease and interventional effects of Chinese herbal medicine].Zhongguo Zhong Yao Za Zhi. 2017 Jul;42(13):2425-2432. doi: 10.19540/j.cnki.cjcmm.20170609.014. Zhongguo Zhong Yao Za Zhi. 2017. PMID: 28840678 Review. Chinese.
-
The interplay among gut microbiota, hypertension and kidney diseases: The role of short-chain fatty acids.Pharmacol Res. 2019 Mar;141:366-377. doi: 10.1016/j.phrs.2019.01.019. Epub 2019 Jan 10. Pharmacol Res. 2019. PMID: 30639376 Review.
Cited by
-
Wnt signaling pathway in spinal cord injury: from mechanisms to potential applications.Front Mol Neurosci. 2024 Jul 24;17:1427054. doi: 10.3389/fnmol.2024.1427054. eCollection 2024. Front Mol Neurosci. 2024. PMID: 39114641 Free PMC article. Review.
-
Pentosan polysulfate alleviates interstitial cystitis/bladder pain syndrome by modulating bile acid metabolism and activating the TGR5 receptor through gut microbiota regulation.Bladder (San Franc). 2025 Mar 24;12(2):e21200036. doi: 10.14440/bladder.2024.0060. eCollection 2025. Bladder (San Franc). 2025. PMID: 40747461 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

