Clinical Significance of a Novel Vasculogenic Mimicry-based Prognostic Model in Hepatocellular Carcinoma
- PMID: 38757323
- PMCID: PMC12376130
- DOI: 10.2174/0109298673298862240510073543
Clinical Significance of a Novel Vasculogenic Mimicry-based Prognostic Model in Hepatocellular Carcinoma
Abstract
Background: Vasculogenic mimicry, a novel neovascularization pattern of aggressive tumors, is associated with poor clinical outcomes.
Objective: The aim of this research was to establish a new model, termed VC score, to predict the prognosis, Tumor Microenvironment (TME) components, and immunotherapeutic response in Hepatocellular Carcinoma (HCC).
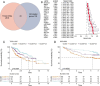
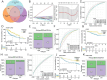
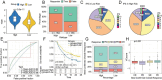
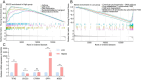
Methods: The expression data of the public databases were used to develop the prognostic model. Consensus clustering was performed to confirm the molecular subtypes with ideal clustering efficacy. The high- and low-risk groups were stratified utilizing the VC score. Various methodologies, including survival analysis, single-sample Gene Set Enrichment Analysis (ssGSEA), Tumor Immune Dysfunction and Exclusion scores (TIDE), Immunophenoscore (IPS), and nomogram, were utilized for verification of the model performance and to characterize the immune status of HCC tissues. GSEA was performed to mine functional pathway information.
Results: The survival and immune characteristics varied between the three molecular subtypes. A five-gene signature (TPX2, CDC20, CFHR4, SPP1, and NQO1) was verified to function as an independent predictive factor for the prognosis of patients with HCC. The high-risk group exhibited lower Overall Survival (OS) rates and higher mortality rates in comparison to the low-risk group. Patients in the low-risk group were predicted to benefit from immune checkpoint inhibitor therapy and exhibit increased sensitivity to immunotherapy. Enrichment analysis revealed that signaling pathways linked to the cell cycle and DNA replication processes exhibited enrichment in the high-risk group.
Conclusion: The VC score holds the potential to establish individualized treatment plans and clinical management strategies for patients with HCC.
Keywords: Hepatocellular carcinoma; immunotherapy.; machine learning; prognostic prediction model; tumor microenvironment; vasculogenic mimicry.
Copyright© Bentham Science Publishers; For any queries, please email at epub@benthamscience.net.
Conflict of interest statement
The authors declare no conflict of interest, financial or otherwise.
Figures



Similar articles
-
A novel vasculogenic mimicry-related nomogram predicts prognosis in hepatocellular carcinoma.Front Genet. 2025 Jul 7;16:1431624. doi: 10.3389/fgene.2025.1431624. eCollection 2025. Front Genet. 2025. PMID: 40692710 Free PMC article.
-
A novel immunogenic cell death-related genes signature for predicting prognosis, immune landscape and immunotherapy effect in hepatocellular carcinoma.J Cancer Res Clin Oncol. 2023 Dec;149(18):16261-16277. doi: 10.1007/s00432-023-05370-1. Epub 2023 Sep 12. J Cancer Res Clin Oncol. 2023. PMID: 37698679 Free PMC article.
-
Anoikis-related lncRNA signature predicts prognosis and is associated with immune infiltration in hepatocellular carcinoma.Anticancer Drugs. 2024 Jun 1;35(5):466-480. doi: 10.1097/CAD.0000000000001589. Epub 2024 Mar 11. Anticancer Drugs. 2024. PMID: 38507233
-
Management of people with early- or very early-stage hepatocellular carcinoma: an attempted network meta-analysis.Cochrane Database Syst Rev. 2017 Mar 28;3(3):CD011650. doi: 10.1002/14651858.CD011650.pub2. Cochrane Database Syst Rev. 2017. PMID: 28351116 Free PMC article.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
Cited by
-
Independent prognostic significance of tissue and circulating microrna biomarkers in hepatocellular carcinoma.Discov Oncol. 2025 Mar 8;16(1):281. doi: 10.1007/s12672-025-02043-y. Discov Oncol. 2025. PMID: 40056315 Free PMC article. Review.
-
Mechanistic link between long noncoding RNA and stability of oncogene reveals stemness and chemoresistance of hepatocellular carcinoma.World J Gastroenterol. 2025 Feb 21;31(7):103400. doi: 10.3748/wjg.v31.i7.103400. World J Gastroenterol. 2025. PMID: 39991677 Free PMC article.
References
-
- Wang Y., Wang J., Li X., Xiong X., Wang J., Zhou Z., Zhu X., Gu Y., Dominissini D., He L., Tian Y., Yi C., Fan Z. N1-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 2021;12(1):6314. doi: 10.1038/s41467-021-26718-6. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

