A PET-Surrogate Signature for the Interrogation of the Metabolic Status of Breast Cancers
- PMID: 38757578
- PMCID: PMC11267279
- DOI: 10.1002/advs.202308255
A PET-Surrogate Signature for the Interrogation of the Metabolic Status of Breast Cancers
Abstract
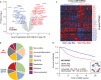
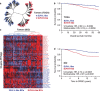
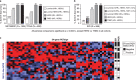
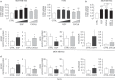
Metabolic alterations in cancers can be exploited for diagnostic, prognostic, and therapeutic purposes. This is exemplified by 18F-fluorodeoxyglucose (FDG)-positron emission tomography (FDG-PET), an imaging tool that relies on enhanced glucose uptake by tumors for diagnosis and staging. By performing transcriptomic analysis of breast cancer (BC) samples from patients stratified by FDG-PET, a 54-gene signature (PETsign) is identified that recapitulates FDG uptake. PETsign is independently prognostic of clinical outcome in luminal BCs, the most common and heterogeneous BC molecular subtype, which requires improved stratification criteria to guide therapeutic decision-making. The prognostic power of PETsign is stable across independent BC cohorts and disease stages including the earliest BC stage, arguing that PETsign is an ab initio metabolic signature. Transcriptomic and metabolomic analysis of BC cells reveals that PETsign predicts enhanced glycolytic dependence and reduced reliance on fatty acid oxidation. Moreover, coamplification of PETsign genes occurs frequently in BC arguing for their causal role in pathogenesis. CXCL8 and EGFR signaling pathways feature strongly in PETsign, and their activation in BC cells causes a shift toward a glycolytic phenotype. Thus, PETsign serves as a molecular surrogate for FDG-PET that could inform clinical management strategies for BC patients.
Keywords: FDG‐PET; breast cancer; gene signature; glycolysis; metabolism.
© 2024 The Authors. Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
A patent application has been submitted for PETsign.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
