Framework for exploring the sensory repertoire of the human gut microbiota
- PMID: 38757952
- PMCID: PMC11237719
- DOI: 10.1128/mbio.01039-24
Framework for exploring the sensory repertoire of the human gut microbiota
Abstract
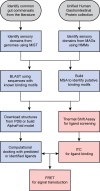
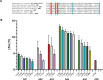
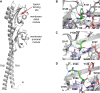
Bacteria sense changes in their environment and transduce signals to adjust their cellular functions accordingly. For this purpose, bacteria employ various sensors feeding into multiple signal transduction pathways. Signal recognition by bacterial sensors is studied mainly in a few model organisms, but advances in genome sequencing and analysis offer new ways of exploring the sensory repertoire of many understudied organisms. The human gut is a natural target of this line of study: it is a nutrient-rich and dynamic environment and is home to thousands of bacterial species whose activities impact human health. Many gut commensals are also poorly studied compared to model organisms and are mainly known through their genome sequences. To begin exploring the signals human gut commensals sense and respond to, we have designed a framework that enables the identification of sensory domains, prediction of signals that they recognize, and experimental verification of these predictions. We validate this framework's functionality by systematically identifying amino acid sensors in selected bacterial genomes and metagenomes, characterizing their amino acid binding properties, and demonstrating their signal transduction potential.IMPORTANCESignal transduction is a central process governing how bacteria sense and respond to their environment. The human gut is a complex environment with many living organisms and fluctuating streams of nutrients. One gut inhabitant, Escherichia coli, is a model organism for studying signal transduction. However, E. coli is not representative of most gut microbes, and signaling pathways in the thousands of other organisms comprising the human gut microbiota remain poorly understood. This work provides a foundation for how to explore signals recognized by these organisms.
Keywords: amino acids; gut microbiome; receptors; signal transduction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
