Parascedosporium putredinis NO1 tailors its secretome for different lignocellulosic substrates
- PMID: 38757984
- PMCID: PMC11218486
- DOI: 10.1128/spectrum.03943-23
Parascedosporium putredinis NO1 tailors its secretome for different lignocellulosic substrates
Abstract
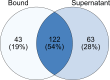
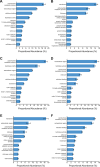
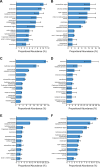
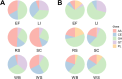
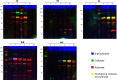
Parascedosporium putredinis NO1 is a plant biomass-degrading ascomycete with a propensity to target the most recalcitrant components of lignocellulose. Here we applied proteomics and activity-based protein profiling (ABPP) to investigate the ability of P. putredinis NO1 to tailor its secretome for growth on different lignocellulosic substrates. Proteomic analysis of soluble and insoluble culture fractions following the growth of P. putredinis NO1 on six lignocellulosic substrates highlights the adaptability of the response of the P. putredinis NO1 secretome to different substrates. Differences in protein abundance profiles were maintained and observed across substrates after bioinformatic filtering of the data to remove intracellular protein contamination to identify the components of the secretome more accurately. These differences across substrates extended to carbohydrate-active enzymes (CAZymes) at both class and family levels. Investigation of abundant activities in the secretomes for each substrate revealed similar variation but also a high abundance of "unknown" proteins in all conditions investigated. Fluorescence-based and chemical proteomic ABPP of secreted cellulases, xylanases, and β-glucosidases applied to secretomes from multiple growth substrates for the first time confirmed highly adaptive time- and substrate-dependent glycoside hydrolase production by this fungus. P. putredinis NO1 is a promising new candidate for the identification of enzymes suited to the degradation of recalcitrant lignocellulosic feedstocks. The investigation of proteomes from the biomass bound and culture supernatant fractions provides a more complete picture of a fungal lignocellulose-degrading response. An in-depth understanding of this varied response will enhance efforts toward the development of tailored enzyme systems for use in biorefining.IMPORTANCEThe ability of the lignocellulose-degrading fungus Parascedosporium putredinis NO1 to tailor its secreted enzymes to different sources of plant biomass was revealed here. Through a combination of proteomic, bioinformatic, and fluorescent labeling techniques, remarkable variation was demonstrated in the secreted enzyme response for this ascomycete when grown on multiple lignocellulosic substrates. The maintenance of this variation over time when exploring hydrolytic polysaccharide-active enzymes through fluorescent labeling, suggests that this variation results from an actively tailored secretome response based on substrate. Understanding the tailored secretomes of wood-degrading fungi, especially from underexplored and poorly represented families, will be important for the development of effective substrate-tailored treatments for the conversion and valorization of lignocellulose.
Keywords: CAZymes; Parascedosporium putredinis NO1; activity-based protein profiling; lignocellulose; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
A bioinformatic workflow for in silico secretome prediction with the lignocellulose degrading ascomycete fungus Parascedosporium putredinis NO1.Mol Microbiol. 2023 Nov;120(5):754-762. doi: 10.1111/mmi.15144. Epub 2023 Aug 30. Mol Microbiol. 2023. PMID: 37646302
-
Whole genome structural predictions reveal hidden diversity in putative oxidative enzymes of the lignocellulose-degrading ascomycete Parascedosporium putredinis NO1.Microbiol Spectr. 2023 Dec 12;11(6):e0103523. doi: 10.1128/spectrum.01035-23. Epub 2023 Oct 9. Microbiol Spectr. 2023. PMID: 37811978 Free PMC article.
-
Comparative exo-proteomics of solid and submerged state fermentation using the lignocellulose degrading Ascomycete Parascedosporium putredinis NO1.Bioresour Technol. 2025 Nov;435:132864. doi: 10.1016/j.biortech.2025.132864. Epub 2025 Jun 21. Bioresour Technol. 2025. PMID: 40550366
-
Proteomic researches for lignocellulose-degrading enzymes: A mini-review.Bioresour Technol. 2018 Oct;265:532-541. doi: 10.1016/j.biortech.2018.05.101. Epub 2018 May 31. Bioresour Technol. 2018. PMID: 29884341 Review.
-
Plant biomass degrading ability of the coprophilic ascomycete fungus Podospora anserina.Biotechnol Adv. 2016 Sep-Oct;34(5):976-983. doi: 10.1016/j.biotechadv.2016.05.010. Epub 2016 Jun 1. Biotechnol Adv. 2016. PMID: 27263000 Review.
Cited by
-
Activity-based probes for dynamic characterisation of polysaccharide-degrading enzymes.Biochem J. 2025 Jul 1;482(13):939-54. doi: 10.1042/BCJ20253060. Biochem J. 2025. PMID: 40591799 Free PMC article. Review.
References
-
- Saha BC, Qureshi N, Kennedy GJ, Cotta MA. 2016. Biological pretreatment of corn stover with white-rot fungus for improved enzymatic hydrolysis. Int Biodeter Biodegr 109:29–35. doi:10.1016/j.ibiod.2015.12.020 - DOI
-
- Salvachúa D, Martínez AT, Tien M, López-Lucendo MF, García F, de Los Ríos V, Martínez MJ, Prieto A. 2013. Differential proteomic analysis of the secretome of Irpex lacteus and other white-rot fungi during wheat straw pretreatment. Biotechnol Biofuels 6:115. doi:10.1186/1754-6834-6-115 - DOI - PMC - PubMed
-
- Cianchetta S, Bregoli L, Galletti S. 2017. Microplate-based evaluation of the sugar yield from giant reed, giant miscanthus and switchgrass after mild chemical pre-treatments and hydrolysis with tailored Trichoderma enzymatic blends. Appl Biochem Biotechnol 183:876–892. doi:10.1007/s12010-017-2470-z - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

