HMGA1 regulates trabectedin sensitivity in advanced soft-tissue sarcoma (STS): A Spanish Group for Research on Sarcomas (GEIS) study
- PMID: 38758230
- PMCID: PMC11101398
- DOI: 10.1007/s00018-024-05250-y
HMGA1 regulates trabectedin sensitivity in advanced soft-tissue sarcoma (STS): A Spanish Group for Research on Sarcomas (GEIS) study
Abstract
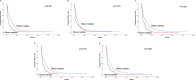
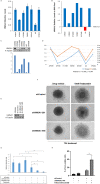
HMGA1 is a structural epigenetic chromatin factor that has been associated with tumor progression and drug resistance. Here, we reported the prognostic/predictive value of HMGA1 for trabectedin in advanced soft-tissue sarcoma (STS) and the effect of inhibiting HMGA1 or the mTOR downstream pathway in trabectedin activity. The prognostic/predictive value of HMGA1 expression was assessed in a cohort of 301 STS patients at mRNA (n = 133) and protein level (n = 272), by HTG EdgeSeq transcriptomics and immunohistochemistry, respectively. The effect of HMGA1 silencing on trabectedin activity and gene expression profiling was measured in leiomyosarcoma cells. The effect of combining mTOR inhibitors with trabectedin was assessed on cell viability in vitro studies, whereas in vivo studies tested the activity of this combination. HMGA1 mRNA and protein expression were significantly associated with worse progression-free survival of trabectedin and worse overall survival in STS. HMGA1 silencing sensitized leiomyosarcoma cells for trabectedin treatment, reducing the spheroid area and increasing cell death. The downregulation of HGMA1 significantly decreased the enrichment of some specific gene sets, including the PI3K/AKT/mTOR pathway. The inhibition of mTOR, sensitized leiomyosarcoma cultures for trabectedin treatment, increasing cell death. In in vivo studies, the combination of rapamycin with trabectedin downregulated HMGA1 expression and stabilized tumor growth of 3-methylcholantrene-induced sarcoma-like models. HMGA1 is an adverse prognostic factor for trabectedin treatment in advanced STS. HMGA1 silencing increases trabectedin efficacy, in part by modulating the mTOR signaling pathway. Trabectedin plus mTOR inhibitors are active in preclinical models of sarcoma, downregulating HMGA1 expression levels and stabilizing tumor growth.
Keywords: HMGA1; Leiomyosarcoma; Soft-tissue sarcoma; Trabectedin; mTOR pathway.
© 2024. The Author(s).
Conflict of interest statement
David S. Moura reports institutional research grants from PharmaMar, Eisai, Immix BioPharma, and Novartis outside the submitted work; travel support from PharmaMar, Eisai, Celgene, Bayer, and Pfizer. Nadia Hindi reports grants, personal fees and non-financial support from PharmaMar, personal fees from Lilly, grants from Eisai, and Novartis, outside the submitted work and research funding for clinical studies (institutional) from PharmaMar, Eli Lilly and Company, AROG, Bayer, Eisai, Lixte, Karyopharm, Deciphera, GSK, Novartis, Blueprint, Nektar, Forma, Amgen and Daichii-Sankyo. Javier Martin-Broto reports research grants from PharmaMar, Eisai, Immix BioPharma, and Novartis outside the submitted work; honoraria for advisory board participation and expert testimony from PharmaMar, Eli Lilly and Company, Bayer, GSK, Novartis, Boehringer Ingelheim, Amgen, Roche, Tecnofarma and Asofarma; and research funding for clinical studies (institutional) from PharmaMar, Eli Lilly and Company, BMS, Pfizer, AROG, Bayer, Eisai, Lixte, Karyopharm, Deciphera, GSK, Celgene, Novartis, Blueprint, Adaptinmune, Nektar, Forma, Amgen, Daichii-Sankyo, Ran Therapeutics, INHIBRX, Ayala Pharmaceuticals, Philogen, Cebiotex, PTC Therapeutics, Inc. and SpringWorks therapeutics. The other authors declare no potential conflicts of interest.
Figures



Similar articles
-
Efficacy of trabectedin in advanced soft tissue sarcoma: beyond lipo- and leiomyosarcoma.Drug Des Devel Ther. 2015 Oct 27;9:5785-91. doi: 10.2147/DDDT.S92395. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 26604682 Free PMC article.
-
Integrated Molecular Characterization of Patient-Derived Models Reveals Therapeutic Strategies for Treating CIC-DUX4 Sarcoma.Cancer Res. 2022 Feb 15;82(4):708-720. doi: 10.1158/0008-5472.CAN-21-1222. Cancer Res. 2022. PMID: 34903601 Free PMC article.
-
Identifying and targeting cancer stem cells in leiomyosarcoma: prognostic impact and role to overcome secondary resistance to PI3K/mTOR inhibition.J Hematol Oncol. 2019 Jan 25;12(1):11. doi: 10.1186/s13045-018-0694-1. J Hematol Oncol. 2019. PMID: 30683135 Free PMC article.
-
Real-world evidence of the efficacy and tolerability of trabectedin in patients with advanced soft-tissue sarcoma.Expert Rev Anticancer Ther. 2020 Nov;20(11):957-963. doi: 10.1080/14737140.2020.1822744. Epub 2020 Sep 29. Expert Rev Anticancer Ther. 2020. PMID: 32930637 Review.
-
The safety and efficacy of trabectedin for the treatment of liposarcoma or leiomyosarcoma.Expert Rev Anticancer Ther. 2016 May;16(5):473-84. doi: 10.1080/14737140.2016.1174582. Epub 2016 Apr 22. Expert Rev Anticancer Ther. 2016. PMID: 27043847 Review.
Cited by
-
Decoding the Epigenome: Comparative Analysis of Uterine Leiomyosarcoma and Leiomyoma.Cancers (Basel). 2025 Aug 9;17(16):2610. doi: 10.3390/cancers17162610. Cancers (Basel). 2025. PMID: 40867239 Free PMC article. Review.
-
Tumor-associated stroma shapes the spatial tumor immune microenvironment of primary Ewing sarcomas.bioRxiv [Preprint]. 2025 Feb 6:2025.01.31.635996. doi: 10.1101/2025.01.31.635996. bioRxiv. 2025. PMID: 39975230 Free PMC article. Preprint.
-
Advancements in Diagnosis and Treatment of Cardiac Sarcomas: A Comprehensive Review.Curr Treat Options Oncol. 2025 Feb;26(2):103-127. doi: 10.1007/s11864-024-01287-0. Epub 2025 Jan 31. Curr Treat Options Oncol. 2025. PMID: 39885109 Review.
References
-
- Demetri GD, et al. Efficacy and safety of trabectedin in patients with advanced or metastatic liposarcoma or leiomyosarcoma after failure of prior anthracyclines and ifosfamide: results of a randomized phase II study of two different schedules. J Clin Oncol. 2009;27(25):4188–4196. doi: 10.1200/JCO.2008.21.0088. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

